ICH Topic Q6B states "When process changes and degradation products result in heterogeneity patterns which differ from those observed in the material used during preclinical and clinical development, the significance of these alterations should be evaluated."
Protein degradation analysis can help customers gain insight into the structure and function of proteins so that they can be better designed and modified to perform specific biological functions. In addition, protein degradation analysis can help people develop new drugs and therapeutic approaches to target protein degradation processes for modulation.
Protein Degradation
Protein catabolism denotes the deterioration of proteins into smaller fragments or free amino acids. This physiological yet vital cellular undertaking facilitates protein equilibrium maintenance, elimination of defective or malformed proteins, and the repurposing of amino acids for neoprotein biosynthesis. The processes of protein degradation predominantly operate via two primary pathways: the ubiquitin-proteasome trajectory and the lysosomal degradation route.
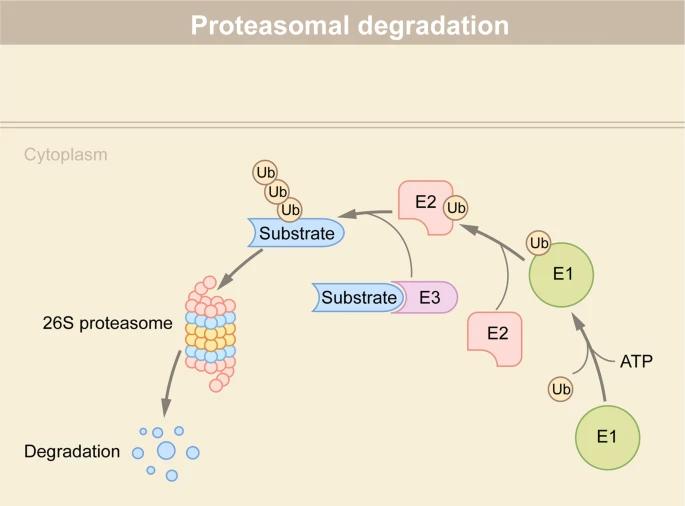 Fig 1. Protein degradation via the ubiquitin-proteasome system (UPS). (Lin Z., et al.; 2022)
Fig 1. Protein degradation via the ubiquitin-proteasome system (UPS). (Lin Z., et al.; 2022)
1. Ubiquitin-proteasome system: This entails the covalent ligation of a diminutive protein, ubiquitin, to proteins designated for catabolism. The ubiquition conjugation onto the target proteins is executed in a sequential pattern, hence constructing a ubiquitin nexus. This chain of ubiquitin molecules serves as a beacon for the proteasome, a voluminous protein aggregate, which recognizes and subsequently disassembles the ubiquitin-labeled protein into truncated peptides, and finally into free amino acids.
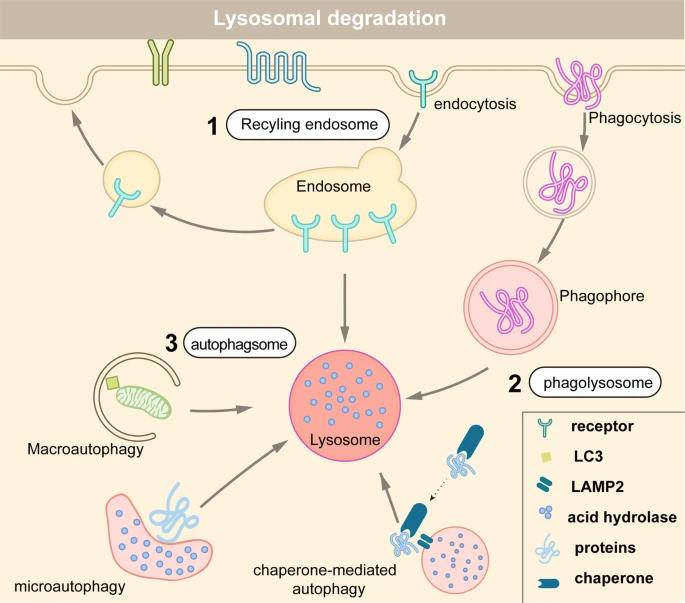 Fig 2. Protein degradation via three distinct lysosome pathways. (Lin Z., et al.; 2022)
Fig 2. Protein degradation via three distinct lysosome pathways. (Lin Z., et al.; 2022)
2. Lysosomal degradation pathway: This trajectory implicates the protein directing towards specialized acidic organelles, the lysosomes. Intrinsically, lysosomes are reservoirs of proteolytic enzymes or proteases, which dismantle proteins into peptides ensuing into free amino acids. Protein ingestion into the lysosomal milieu may occur via various routes, encompassing chaperone-facilitated autophagy, microautophagy, and macroautophagy.
Protein Degradation as a Therapy for Disease

Selective or specific degraders: These are molecular structures that bind to proteins associated with diseases and induce their degradation through the ubiquitin-proteasome system or alternative degradation pathways. Therapies influenced by protein degradation hold considerable promise to magnify selectivity, augment efficacy, and lower toxicity levels in comparison to conventional drugs that target specific proteins. Nevertheless, whilst the potential is significant, numerous challenges persist in the development of highly selective degraders, such as understanding the potential for off-target effects and optimizing delivery methods.
Protein Degradation Detection
Western blotting: This approach facilitates the identification and quantification of specific proteins within a given sample, utilizing the specificity of antibodies. Through juxtaposition of protein concentrations in untreated and treated samples, inferences can be made regarding potential protein degradation.
Pulse-chase experiments: This technique poses a collaborative utility of radioactive or fluorescent tags acting as labels for proteins and subsequently monitoring their rate of degradation over a specified timeframe. The rate of degradation can be deduced by tracking the decrement in the signal from labeled proteins.
Proteomics: With the adoption of Mass spectrometry-based proteomic methodologies, one can detect alterations in protein levels, identify fragmented proteins that have undergone degradation, and postulate protein degradation rates in a manner that permits high-throughput analysis.
Immunofluorescence microscopy: This technique permits the visualization and determination of abundance and localization of particular proteins within cellular structures. Variations in the concentration or subcellular positioning of proteins can deliver valuable insights into the processes and mechanisms of protein degradation.
Service Process
Our Goals
Creative Proteomics is committed to helping our customers with their protein degradation analysis needs. We offer services related to protein drug characterization, providing you with fast, quality answers to support your research needs. If you are interested in our services or have any questions, please contact us; we welcome your feedback and look forward to working with you.
Reference
- Lin Z., et al.; Targeted protein degradation: mechanisms, strategies and application. Signal Transduction and Targeted Therapy. 2022, 7(1):113.
Related Sections
Services
Applications
For research use only, not intended for any clinical use.

 Fig 1. Protein degradation via the ubiquitin-proteasome system (UPS). (Lin Z., et al.; 2022)
Fig 1. Protein degradation via the ubiquitin-proteasome system (UPS). (Lin Z., et al.; 2022) Fig 2. Protein degradation via three distinct lysosome pathways. (Lin Z., et al.; 2022)
Fig 2. Protein degradation via three distinct lysosome pathways. (Lin Z., et al.; 2022)

