Introduction to Interferon (IFN)
Interferon (IFN) is a classification of signaling proteins synthesized and secreted by host cells in reaction to the detection of various pathogens, inclusive of viruses, bacteria, parasites, and oncogenic cells. This collective unit of signaling proteins comprises Type I, II and III interferons.
Type I interferon is predominately generated by leukocytes and fibroblasts, the latter being a cellular constituent of connective tissues. These types are typically produced in consequence to a viral invasion. Functionally, type I interferon instigates an augmentation of anti-viral defenses within peripheral cells. IFN-γ disseminated by innate natural killer cells and selective clusters of T-cells, respectively, constitutes the singular Type II interferon. Upon recognition of virally-infected or malignant cells, this type is emitted, thereby strengthening the overall immune responses.
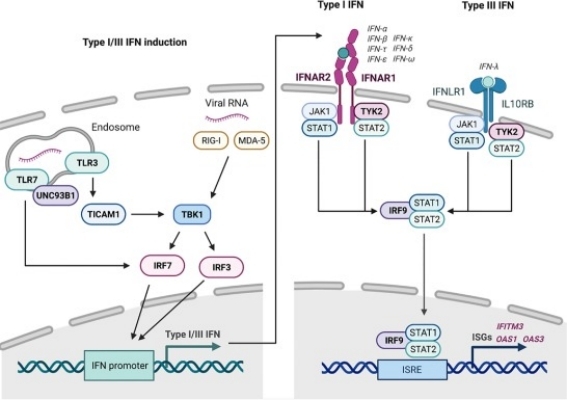 Fig 1. Overview of the type I and type III interferon (IFN) responses. (Stertz, Silke, and Benjamin G. Hale. 2021)
Fig 1. Overview of the type I and type III interferon (IFN) responses. (Stertz, Silke, and Benjamin G. Hale. 2021)
Type III interferon, designated as IFN-λs, exhibits functional similarities to Type I, but notable differences exist, primarily, its diminished inflammatory potential. This demarcated division of signaling proteins plays a pivotal role in host defense against a myriad of pathogenic threats.
Introduction to IFNs Structure
IFN-α constitutes a single polypeptide chain, typically comprising 165 amino acids. Mirroring the topological organization seen in other cytokines, it displays a 3D architectural structure characterized by four α-helices, arranged anti-parallel, that formulate a uniquely longer-than-wide bundle. This helical cluster includes a subtle bend present within the second helix.
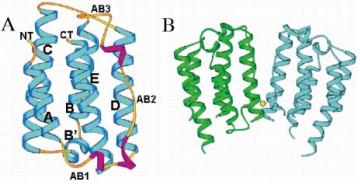 Fig 2. Molecular structure of IFN-α2b. (Nagabhushan, T. L., et al. 2002)
Fig 2. Molecular structure of IFN-α2b. (Nagabhushan, T. L., et al. 2002)
IFN-α's amino acid sequence possesses six cysteine residues, four of which contribute to two intramolecular disulfide bridges, a key factor in maintaining its 3D conformational structure and ensuring the molecule's stability. The residual two cysteines exist in an unpaired state, a condition hypothesized to foster formation of intermolecular disulfide bridges, and thus contributing to the manifestation of multiple functional forms of IFN-α within the organism.
IFN-β protein, contingent upon its subtype and species of origin, has been observed to be composed of approximately 166 to 167 amino acids. This marginal variation is attributable to the protein's capacity to undergo post-translational modifications. The molecular weight of IFN-β fluctuates within a range of 20 to 25 kilodaltons (kDa).
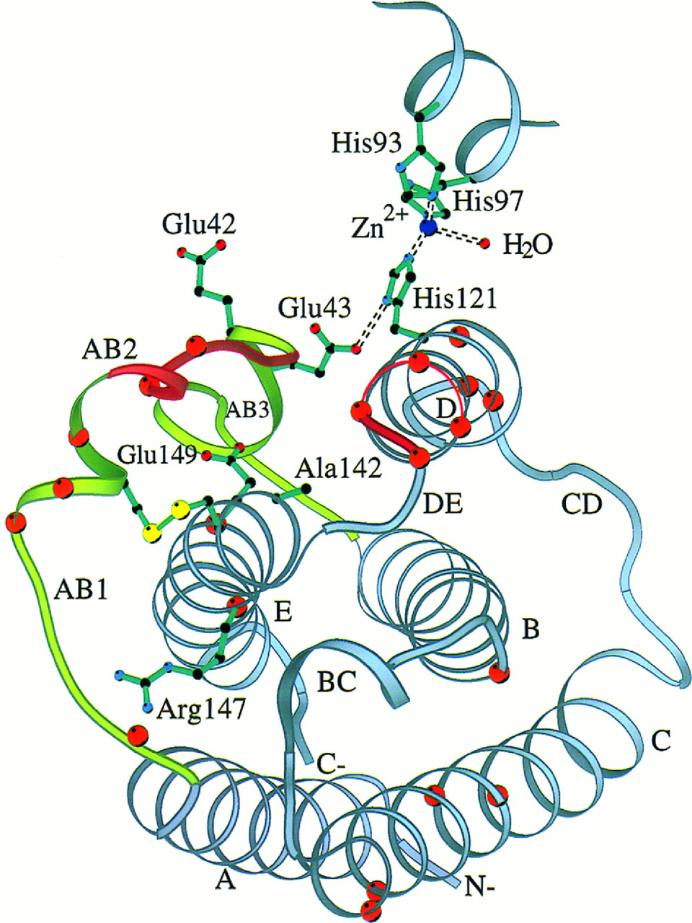 Fig 3. Ribbon diagram of huIFN-β Cα backbone with side chains of residues known to be important for activity. (Karpusas, Michael, et al. 1997)
Fig 3. Ribbon diagram of huIFN-β Cα backbone with side chains of residues known to be important for activity. (Karpusas, Michael, et al. 1997)
In terms of its tertiary structure, the IFN-β protein is predominantly arrayed with several alpha helices, with a noted absence of beta sheets. Interconnected by loops, the stability of this overall configuration is maintained via the presence of disulfide bonds. A salient characteristic feature is the incorporation of an archetypical 'protein fold' within the molecular structure of IFN-β, a structural attribute common throughout the entire interferon family.
Each individual protein chain, or monomer, that constitutes IFN-γ, comprises approximately 143 amino acids and possesses a molecular weight roughly around 25 kDa. The positioning of these monomers is characterized by an antiparallel orientation, signifying that the monomers are juxtaposed with their respective directionalities opposing each other. The significance of this unique configuration lies in its essential role in facilitating the successful binding of IFN-γ with its receptor.
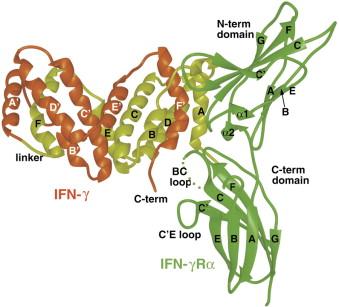 Fig 4. The Structure and Activity of a Monomeric Interferon-γ:α-Chain Receptor Signaling Complex. (Randal, Michael, and Anthony A. Kossiakoff. 2001)
Fig 4. The Structure and Activity of a Monomeric Interferon-γ:α-Chain Receptor Signaling Complex. (Randal, Michael, and Anthony A. Kossiakoff. 2001)
Furthermore, the binding locus of IFN-γ with its corresponding receptor (the subunits IFNGR1 and IFNGR2) necessitates the dimerization of both chains, incorporating amino acids situated in both the N-terminus and the C-terminus - terms referring to the initial and terminal ends of the protein chain, respectively.
What Can We Offer?
Why Choose Our Service

Expertise

Collaborative Approach

Cutting-Edge Technology
Renowned for our forefront position within the biotechnology domain, herein Creative Proteomics, we present a plethora of services specifically designed to tackle complex challenges in protein pharmaceutical characterization. Should you need further clarification or seek more information, we invite you to establish a conversation or make enquiries with us.
References
- Stertz, Silke, and Benjamin G. Hale. Interferon system deficiencies exacerbating severe pandemic virus infections. Trends in microbiology. 2021, 29.11: 973-982.
- Nagabhushan, T. L., et al. Type I interferon structures: Possible scaffolds for the interferon-alpha receptor complex. Canadian Journal of Chemistry. 2002, 80(8), 1166–1173.
- Karpusas, Michael, et al. The crystal structure of human interferon β at 2.2-Å resolution. Proceedings of the National Academy of Sciences. 1997, 94.22: 11813-11818.
- Randal, Michael, and Anthony A. Kossiakoff. The structure and activity of a monomeric interferon-γ: α-chain receptor signaling complex. Structure. 2001, 9.2: 155-163.
Related Sections
Services
Applications
Creative Proteomics specializes in protein drug characterization, and we offer a range of services to help our clients understand and optimize their protein drug products. Our services include, but are not limited to:
|
Protein Drug characterization
|
Reaearch Project
|
Method
|
Application
|
|
Protein Structure Confirmation Service
|
Primary Structure Analysis
|
X-ray crystal diffraction,
nuclear magnetic resonance (NMR) spectroscopy,
ellman's assay,
ion exchange chromatography (IEC),
edman degradation,
mass spectrometry (MS), etc.
|
Protein functions, disease mechanisms, and drug design, etc.
|
|
Higher-Order Structure Analysis
|
|
Post-Translational Modification (PTM) Analysis Service
|
Protein Glycan Analysis
|
Mass spectrometry (MS),
nuclear magnetic resonance (NMR) spectroscopy,
lectin affinity chromatography,
liquid chromatography-mass spectrometry (LC-MS), etc.
|
Disease mechanism research, drug discovery and development, regulation of biological processes, clinical diagnostic, bioinformatics, etc.
|
|
Protein Acetylation Analysis
|
|
Protein Phosphorylation Analysis
|
|
Protein Ubiquitination Analysis
|
|
Protein Deamidation Analysis
|
|
Protein Oxidation Analysis
|
|
Protein Methylation Analysis
|
|
Protein Alkylation Analysis
|
|
Protein Sulfation Analysis
|
|
Proteolysis Analysis
|
|
Protein Truncation Analysis
|
|
Protein Physicochemical Property Determination Service
|
Isoelectric Point (PI) Determination
|
Isoelectric focusing (IEF),
thioflavin T (ThT) fluorescence assay,
western blotting,
ultraviolet-visible spectrometry,
fluorescence spectrometry, etc.
|
Protein structure study, protein interaction study, protein modification study, protein purity and quantitative analysis, clinical diagnosis, etc.
|
|
Charge Variant Analysis
|
|
Extinction Coefficient Determination
|
|
Protein Aggregation Analysis
|
|
Protein Degradation Analysis
|
|
Thermal (Tm) Stability Analysis
|
|
Protein Quantitation
|
|
Protein Purity Service
|
/
|
Composition-based and activity-based analyses,
mass spectrometry (MS),
high performance liquid chromatography,
capillary electrophoresis, etc.
|
Biomedical research, biological research, clinical diagnostics, etc.
|
|
Protein Impurities Service
|
Host Cell Protein (HCP) Analysis
|
High performance liquid chromatography (HPLC),
mass spectrometry (MS),
size-exclusion chromatography (SEC),
capillary electrophoresis, etc.
|
Quality control, safety assessment, clinical diagnostics, bioprocess research, etc.
|
|
Residual Host Cell DNA (HCD) Analysis
|
|
Residual Protein A Analysis
|
|
Process Related Impurities and Residual Analysis
|
|
Protein Biosafety Analysis Service
|
Bacterial Endotoxins Testing
|
Limulus amebocyte lysate (LAL),
membrane filtration,
PCR,
nucleic acid testing (NAT),
massively parallel sequencing (MPS), etc.
|
Biomedical research, food safety testing, environmental science, industrial production, etc.
|
| Bioburden Testing |
| Sterility Testing |
| Abnormal Toxicity Testing |
| Mycoplasma Testing |
| Subvisible Particles Analysis |
| Visible Particles Analysis |
For research use only, not intended for any clinical use.

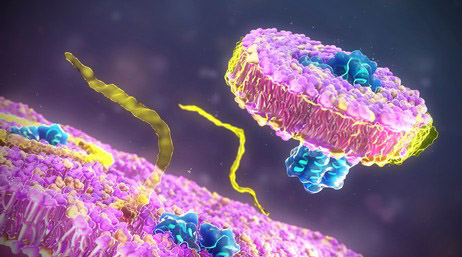
 Fig 1. Overview of the type I and type III interferon (IFN) responses. (Stertz, Silke, and Benjamin G. Hale. 2021)
Fig 1. Overview of the type I and type III interferon (IFN) responses. (Stertz, Silke, and Benjamin G. Hale. 2021) Fig 2. Molecular structure of IFN-α2b. (Nagabhushan, T. L., et al. 2002)
Fig 2. Molecular structure of IFN-α2b. (Nagabhushan, T. L., et al. 2002) Fig 3. Ribbon diagram of huIFN-β Cα backbone with side chains of residues known to be important for activity. (Karpusas, Michael, et al. 1997)
Fig 3. Ribbon diagram of huIFN-β Cα backbone with side chains of residues known to be important for activity. (Karpusas, Michael, et al. 1997) Fig 4. The Structure and Activity of a Monomeric Interferon-γ:α-Chain Receptor Signaling Complex. (Randal, Michael, and Anthony A. Kossiakoff. 2001)
Fig 4. The Structure and Activity of a Monomeric Interferon-γ:α-Chain Receptor Signaling Complex. (Randal, Michael, and Anthony A. Kossiakoff. 2001)


