Quantitative analysis of proteins provides insight into protein function, interactions and expression levels, which is important for biological research and disease diagnosis, etc. Creative Proteomics provides protein quantification services to meet the requirements of ICH Q6B guidelines for characterization of physicochemical properties of biological products.
Protein Quantitation
The technique of calculating the concentration or amount of protein in a sample is known as protein quantification. Because it can provide critical details about their function, abundance, and role in various research, clinical, and technological applications, the concentration of proteins in a sample is a critical step in many of these applications.
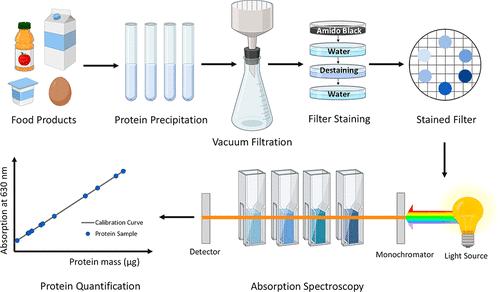 Fig 1. Protein Quantification in Complex Matrices. (Cristiana V. R., et al.; 2022)
Fig 1. Protein Quantification in Complex Matrices. (Cristiana V. R., et al.; 2022)
In several disciplines such as biochemistry, molecular biology, proteomics and drug development, protein quantification is critical. It can be used to evaluate protein concentrations in different samples or experimental settings, assess protein purity and quality, measure protein expression levels, and identify protein-protein interactions.
Workflow of Protein Quantitation
The method of choice for determining a protein's or peptide's concentration in a solution depends on a variety of variables, and the majority of techniques call for soluble analytes like proteins, peptides, post-translationally modified proteins (like glycosylation), or chemically modified proteins (like polyethylene glocalization), among others. The process of choosing the best assay is shown in the flowchart in Figure 2 for common protein purification processes.
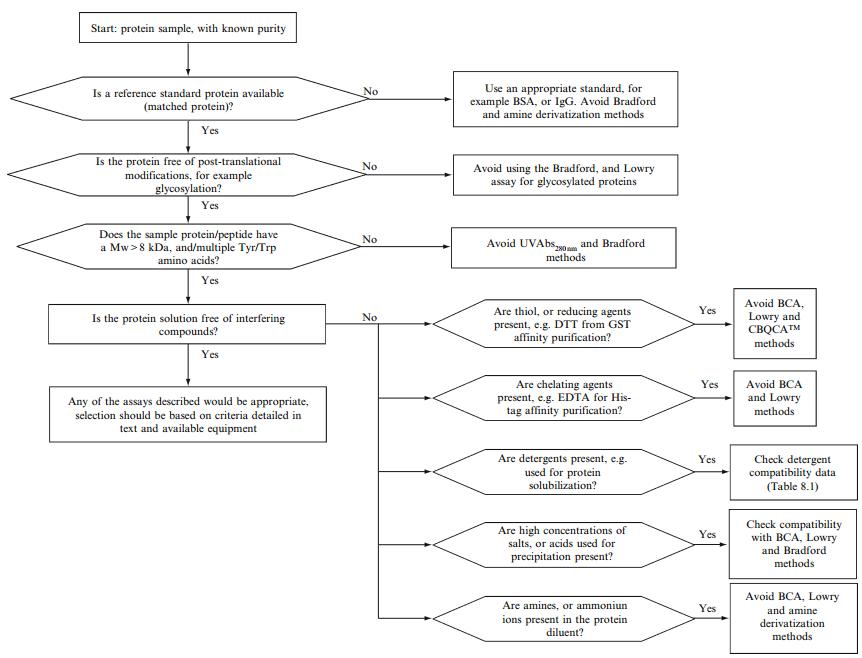 Fig 2. Flow chart for the selection of assays for quantitation or proteins in common protein purification procedures. (Noble, J. E., & Bailey, M. J. A. 2009)
Fig 2. Flow chart for the selection of assays for quantitation or proteins in common protein purification procedures. (Noble, J. E., & Bailey, M. J. A. 2009)
Protein Quantitation Methodology
1. Ultraviolet Absorption Spectroscopy
1.1 Ultraviolet absorbance at 280 nm (Range: 20–3000 μg)

Tyrosine and tryptophan, two aromatic amino acids, dominate the UV absorption spectra that is typical of proteins near 280 nm. This approach will produce inaccurate results if the main sequence does not contain any of these amino acids or only contains them in trace amounts. A protein-free sample buffer solution should be used to analyze a "blank" sample instead of buffer components with high UV absorbance, including certain detergents, particularly Triton X-100. Although UV absorbance is frequently employed to measure protein concentration, the Beer-Lambert rule can be applied to correctly quantify the protein content if the protein's molar extinction coefficient is known.
1.2 Ultraviolet absorbance at 205 nm (Range: 1–100 μg)

Maximum photon absorption by peptide bonds occurs below 210 nm. However, measurements can be done at far longer wavelengths due to the large absorption peaks of peptide bonds, which offers numerous advantages in terms of apparatus and measurement accuracy. Due to interference from solvent and biological buffer components, absorbance at 214 and 220 nm is frequently employed to quantify proteins and peptides. Measurements at Abs205 nm are more sensitive and less varied amongst proteins than measurements at Abs280 nm because proteins contain a lot of peptide bonds.
2. Coomassie Blue (Bradford) Protein Assay (Range: 1–50 μg)

The Coomassie Blue (Bradford) Protein Assay is a protein assay that uses dye to measure the concentration of proteins in samples. In this test, an acidic solution is used to bind proteins with the Coomassie Brilliant Blue dye. The dye changes from brown to blue as a result of the protein it binds to, changing the dye's absorption spectrum. The amount of protein in the sample is directly correlated with the intensity of the blue hue.
3. Lowry (Alkaline Copper Reduction Assays) (Range: 5–100 μg)
The Lowry protein assay is a biochemical assay for determining the total level of protein in a solution. The test is based on the reaction of copper ions with proteins in an alkaline (basic) solution, followed by reduction of the resultant complex by the Folin-Ciocalteu reagent under acidic conditions. The amount of reduced Folin-Ciocalteu reagent is proportional to the protein concentration, which can be calculated by measuring the absorbance of the solution at 750 nm. The Lowry assay is sensitive and can measure protein concentrations as low as 5-10 µg/ml, making it widely used in biochemical studies. It is, however, affected by the presence of reducing agents, detergents, and other substances, hence samples need to be purified before assay.
4. Bicinchoninic Acid (BCA) (Range: 0.2–50 μg)

Bicinchoninic Acid, commonly abbreviated as BCA, plays an integral role in biochemistry as it is extensively applied in the quantification of protein concentration present in a solution. Utilizing the principles of a colorimetric assay, the BCA Protein Assay produces a color change to signify the presence and concentration of a protein-specific substance. This method, also known as the Smith Assay, offers advantages over other methods, such as the Lowry method, in terms of sensitivity and decreased susceptibility to interference from diverse chemicals. Despite these advantages, the BCA assay is unable to differentiate among various protein types necessitating the use of complementary methods for comprehensive protein analysis.
5. Amine Derivatization (Range: 0.05–25 μg)

Amine modification represents a bio-analytical technique associated with altering the structure of an amine molecule within a protein. This process is conventionally aimed at augmenting the visibility or assessability of a protein. To achieve this, a chemical reagent of choice is employed to induce reactions with either primary or secondary amines, leading to the formation of a chemically-transformed entity. This modified molecule can then be detected and scrutinized through spectrophotometric methods or other sophisticated analytical methodologies one might have at their disposal.
6. Detergent-Based Fluorescent Detection (Range: 0.02–2 μg)

The fluorescent detergent-based detection methodology employs luminescent dyes conjugated to detergent molecules for the identification and quantification of proteins. This technique is premised on the principle of a non-covalent interaction established between the protein entity and the detergent molecule, subsequently adorned with a luminous dye. This binding mechanism triggers a conformational transformation within the protein-detergent complex, resulting in an amplified fluorescence output. The subsequent quantification of the protein of interest is achieved by comparing the resultant fluorescence signal, derived from the aforementioned dye-detergent-protein tri-component complex, with a predetermined reference standard curve constructed via the known concentrations of a reference protein, using fluorescence spectrophotometry. This comparison enables the accurate determination of protein concentration with this innovative fluorescent detergent-based detection approach.
Why Us?

Our Goals
As a highly-regarded expert in the field of biochemistry with a specific focus on proteomics, I am dedicated to sharing how Creative Proteomics approaches protein quantification services. Our mission is to provide a precise and trustworthy protein enumeration methodology for fellow scientists involved in diverse sectors of research. We primarily focus on providing an array of protein quantification techniques capable of meeting the unique needs of our clientele. Our methodologies span from conventional dye-based assays such as Bradford, Lowry, and BCA, to more innovative assays.
At Creative Proteomics, we ensure diligent execution of high-quality analytical processes, swift delivery periods, and the generation of exact quantitative protein data that will assist in moulding your research. I encourage fellow researchers interested in leveraging our expert protein quantification services for their work to reach out to us.
Reference
- Cristiana V. R., et al.; Protein Quantification in Complex Matrices. Journal of Chemical Education. 2022, 99, 3, 1488–1496.
Related Sections
Services
Applications
For research use only, not intended for any clinical use.

 Fig 1. Protein Quantification in Complex Matrices. (Cristiana V. R., et al.; 2022)
Fig 1. Protein Quantification in Complex Matrices. (Cristiana V. R., et al.; 2022) Fig 2. Flow chart for the selection of assays for quantitation or proteins in common protein purification procedures. (Noble, J. E., & Bailey, M. J. A. 2009)
Fig 2. Flow chart for the selection of assays for quantitation or proteins in common protein purification procedures. (Noble, J. E., & Bailey, M. J. A. 2009)







