ICH Topic Q6B states "Process-related impurities encompass those that are derived from the manufacturing process, i.e., cell substrates (e.g., host cell proteins, host cell DNA), cell culture (e.g., inducers, antibiotics, or media components), or downstream processing."
As a foremost specialist in proteomics and genomics, Creative Proteomics offers in-depth analysis services for detecting and quantifying residual host cell DNA in biological products, crucial for quality control in biopharmaceuticals, vaccines, and gene therapy development.
Background
Post-production residuals of HCD refer to the minute fragments of DNA left behind from the host organism exploited in the generation of biological products, which include, but are not limited to, vaccines and therapeutic proteins. Throughout the production protocol, host cells—commonly bacterial or mammalian—are employed to express the targeted product. Although rigorous purification processes are undertaken, negligible quantities of this host cell DNA may be persistent in the final output, annotated as residual Host Cell DNA.
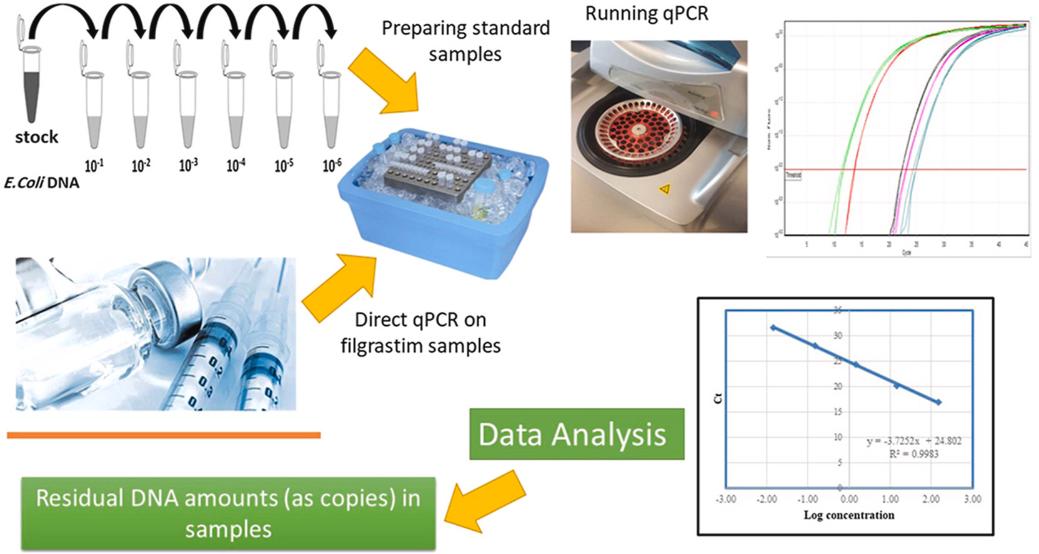 Fig 1. Direct quantitative detection of host cell residual DNA in recombinant Filgrastim by qPCR. (Gholizadeh-Hashjin, A., et al.; 2021)
Fig 1. Direct quantitative detection of host cell residual DNA in recombinant Filgrastim by qPCR. (Gholizadeh-Hashjin, A., et al.; 2021)
Risk Assessment
Virus Infectivity
A virus is incapable of propagating independent of a specific host cell. Once it infiltrates a host organism, the virus capitalizes on cellular machinery for the purpose of replication and continued dissemination. The process commences with viral infiltration of a cell, during which it introduces its own genetic code - either DNA or RNA - into the cell's nucleus. In certain scenarios, the viral genetic code can physically embed itself into the DNA of the host cell, an event noted as integration. Following integration, the cell can continue its viral production independent of new viral infiltrations. This capacity often results in chronic or latent infections, where the viral presence can remain inactive or dormant for an extended span before reactivating.
Oncogenicity
Within the sphere of cellular biology, viruses have the aptitude to infiltrate host cells and intermingle their genetic material with the host's DNA, a phenomenon categorized as DNA integration. Following this assimilation, the recombinant viral DNA can instigate mutations within the host's genomic sequence which has the capacity to devolve into carcinogenesis. A subset of these viruses harbor oncogenes, genetic material inherently predisposed to induce cancer. This integration of oncogenes into the host cell DNA may catalyze unchecked cellular proliferation and division, setting the stage for tumorigenesis.
Immunogenicity

The issue of host cell DNA inducing immunogenicity becomes particularly salient within the contexts of gene therapy or the delivery of recombinant DNA. In these scenarios, the host cell DNA may undergo integration into biopharmaceutical products, potentially rousing an immune response. This immune recognition can occur due to specific sequences or components within the DNA that the host's immune system can identify. Such a response can escalate into a substantial problem, particularly if the DNA sequence mirrors or shares identity with a sequence located within a human autoantigen. This can potentially activate an autoimmune reaction—a detrimental misfiring of the body's immune machinery against its own proteins.
Methods for Residual Host Cell DNA (HCD) Analysis
1. Quantitative PCR (qPCR)
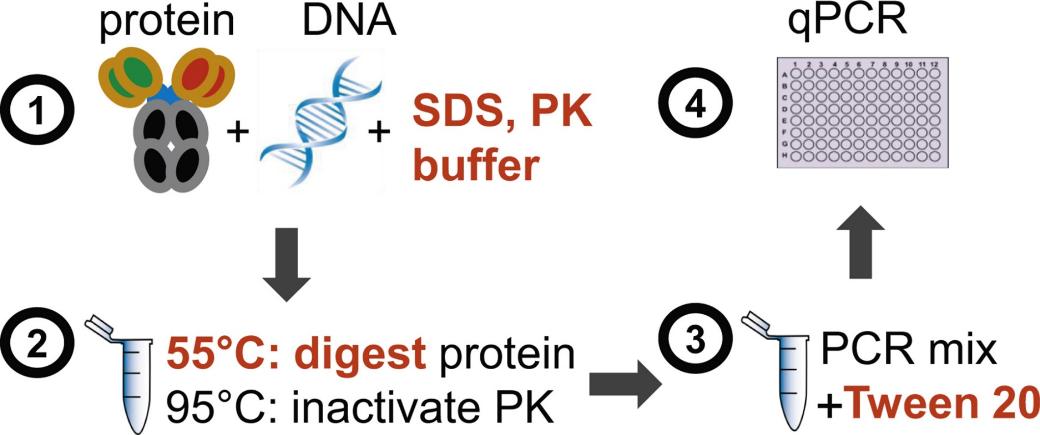 Fig 2. Direct real-time quantitative PCR for measurement of host-cell residual DNA in therapeutic proteins. (Peper, G., et al.; 2014)
Fig 2. Direct real-time quantitative PCR for measurement of host-cell residual DNA in therapeutic proteins. (Peper, G., et al.; 2014)
The Polymerase Chain Reaction (PCR) method is widely recognized as one of the principle techniques employed in the detection of residual HCD. Using Quantitative PCR (qPCR) to test Residual Host Cell DNA (HCD) involves several steps: DNA extraction, DNase treatment, preparing qPCR reactions, establishing quantification standards & controls, thermocycling conditions, data analysis:
2. Capillary Gel Electrophoresis
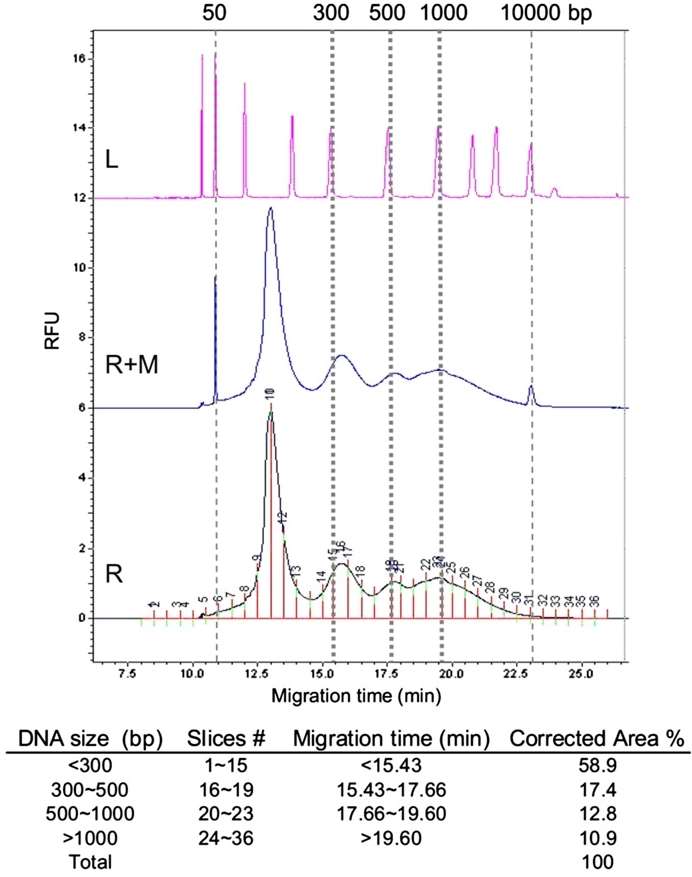 Fig 3. Demonstration of CGE data analysis for determination of DNA size distribution. (Shen, X., et al.; 2013)
Fig 3. Demonstration of CGE data analysis for determination of DNA size distribution. (Shen, X., et al.; 2013)
The Capillary Gel Electrophoresis (CGE) is an excellent method for checking for residual host cell DNA in situations where the purity of a biological sample is essential. In medical and scientific research, contamination of samples can lead to misleading or inaccurate findings. By employing the CGE technique, remaining DNA segments from the host cell can be effectively identified and quantified, ensuring the validity of experimental results.
Why Choose Us?

Our Goals
Creative Proteomics is committed to the precise identification and accurate quantification of residual HCD that may be present in biopharmaceutical samples. Our focus is on achieving superior sensitivity levels in HCD analysis, which allows for the detection and quantification of even low-abundance HCD. We also strive to define the limit of detection in line with pertinent regulatory standards. If you require further information or wish to discuss specific requirements, we invite you to get in touch with us.
References
- Gholizadeh-Hashjin, A., et al.; Direct quantitative detection of host cell residual DNA in recombinant Filgrastim by qPCR. Analytical Biochemistry. 2021, 629, 114296.
- Peper, G., et al.; Direct real-time quantitative PCR for measurement of host-cell residual DNA in therapeutic proteins. Journal of Pharmaceutical and Biomedical Analysis. 2014, 100, 123–130.
- Shen, X., et al.; Size analysis of residual host cell DNA in cell culture-produced vaccines by capillary gel electrophoresis. Biologicals. 2013, 41(3), 201–208.
Related Sections
Services
Applications
For research use only, not intended for any clinical use.

 Fig 1. Direct quantitative detection of host cell residual DNA in recombinant Filgrastim by qPCR. (Gholizadeh-Hashjin, A., et al.; 2021)
Fig 1. Direct quantitative detection of host cell residual DNA in recombinant Filgrastim by qPCR. (Gholizadeh-Hashjin, A., et al.; 2021)


 Fig 2. Direct real-time quantitative PCR for measurement of host-cell residual DNA in therapeutic proteins. (Peper, G., et al.; 2014)
Fig 2. Direct real-time quantitative PCR for measurement of host-cell residual DNA in therapeutic proteins. (Peper, G., et al.; 2014) Fig 3. Demonstration of CGE data analysis for determination of DNA size distribution. (Shen, X., et al.; 2013)
Fig 3. Demonstration of CGE data analysis for determination of DNA size distribution. (Shen, X., et al.; 2013)
