ICH Q6B states "Characterisation of a biotechnological or biological product (which includes the determination of physicochemical properties, biological activity, immunochemical properties, purity and impurities) by appropriate techniques is necessary to allow relevant specifications to be established."
Creative Proteomics, a leading company in the field of proteomics research, places its vast expertise and advanced technologies at the disposal of customers by offering them highly precise and reliable protein purity determination services. We ensure that every procedure is carried out meticulously, ensuring that the results are accurate and reliable, which can significantly contribute to the progress and success of their customers' research projects.
Overview
Protein purity defines the extent to which a particular protein of interest has been successfully separated and isolated from any contaminating proteins or other components within a given sample. Ensuring a high degree of protein purity is a cornerstone of research in several areas of biological sciences, spanning from biotechnology to molecular biology, biochemistry, and medical research. The reliance on proteins of high purity in these fields is due to the necessity for rigorous and reliable experimental results. Profiling protein purity is generally achieved via quintessential methodologies like SDS-PAGE, chromatography, and mass spectrometry.
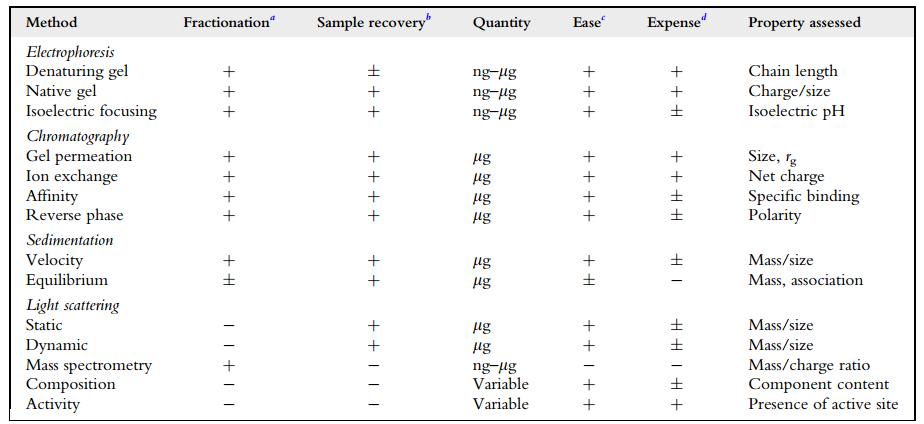 Fig 1. Determination of Protein Purity. (Rhodes, D. G., & Laue, T. M.; 2009)
Fig 1. Determination of Protein Purity. (Rhodes, D. G., & Laue, T. M.; 2009)
Prerequisites for Protein Purity Determination
Evaluating the purity of a protein sample is not a straightforward operation. It invariably necessitates the identification of particular contaminants to assert the purity of the protein mixture. This assessment is pivotal, whether it underpins the interpretation of analytical output, reveals, or authenticates the safety of a biopharmaceutical product.
The initial step in appraising the purity of a sample is the identification of the type of impurity that we aim to quantify. This could be nucleic acids, carbohydrates, lipids, unrelated proteins, isoenzymes, or inactive enzymes. Following this, we need to define a distinctive attribute (be it chemical, analytical, or physical) of the assumed contaminant in the context of the relevant proteins under specific solution conditions.
Purity representation is essentially an assertion that the level of contamination is lower than a pre-established threshold. It is important to note here that it is not necessary to fully describe the contaminant. Even after reducing the concentration of a potential contaminant to below the detection limit of a chromatographic method, the exact nature of the contaminant may remain unidentified. It should be emphasized that the level of detectability depends on the sensitivity of the chosen detection methodology.
Protein Purity Determination Mehodology
1. Composition-Based and Activity-Based Analyses
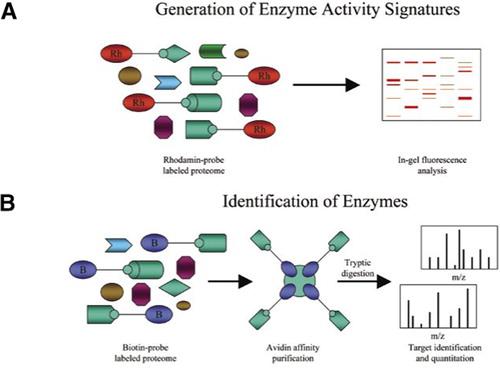 Fig 2. Combination of activity-based protein profiling (ABPP) and mass spectrometry (MS)based proteomic profiling to identify specific activities within complex human samples. (Conrads, T. P., et al.; 2006)
Fig 2. Combination of activity-based protein profiling (ABPP) and mass spectrometry (MS)based proteomic profiling to identify specific activities within complex human samples. (Conrads, T. P., et al.; 2006)
Through assessing the molecular weight of distinct amino acids, repair assemblies, or operational regions, one can establish the purity level of a particular sample. It may be practicable to compute the unit activity as an index of relative purity in instances where the specific activity of an unadulterated chemical compound is established. The determination of known activity or specific analyte in a sample and the mass of the protein (or other material) used for analysis are generally the two main readings required. The mass of the substance that is subjected to analysis is then employed to calculate the prospective amount of analyte. The ratio between the actual quantified value and the predicted amount is then employed as a gauge to denote purity.
2. Electrophoretic Methods
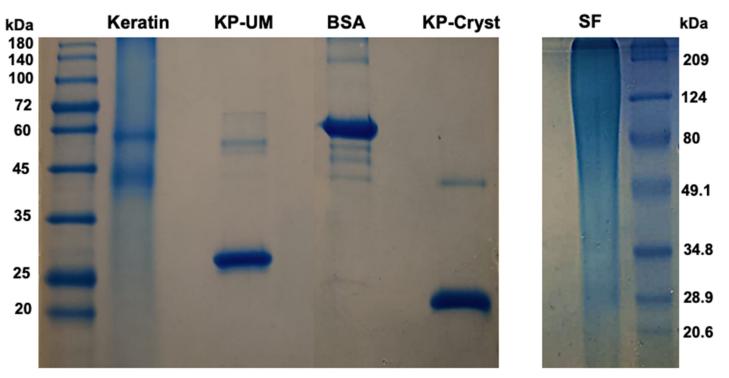 Fig 3. Protein purity analysis by SDS-PAGE in a 12.5% polyacrylamide gel. (Tinoco, A., et al.; 2021)
Fig 3. Protein purity analysis by SDS-PAGE in a 12.5% polyacrylamide gel. (Tinoco, A., et al.; 2021)
Upon subjecting a specific protein sample to electrophoresis, the protein will move at a velocity that is dictated by three factors: its size, its charge, and its conformational shape. This implies that proteins of varying kind will proceed at different rates, facilitating their separation. In an optimized scenario, if a protein sample is pure, it should represent only a single kind of protein. This, in turn, should depict just one band or spot on the electrophoresis gel. Observation of more than one band or spot is indicative of contamination. It signals the presence of diverse types of protein in the sample, thus confirming that the sample lacks purity.
3. Chromatographic Methods
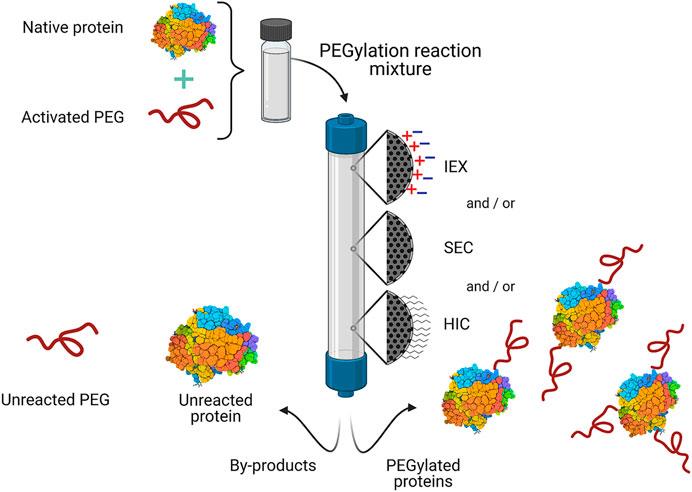 Fig 4. General scheme for the purification of commercial PEGylated proteins by chromatography. (Calef, S.T., et al.; 2021)
Fig 4. General scheme for the purification of commercial PEGylated proteins by chromatography. (Calef, S.T., et al.; 2021)
Chromatographic detection is a sophisticated method extensively employed in the realm of biology for the identification and quantification of various constituents within a complex matrix. This technique plays a crucial role in assessing the purity of proteins; it achieves this by resolving the proteins of interest from the myriad contaminants present in a given sample. Enhanced separation of proteins from impurities invariably augurs an elevated level of purity. Consequently, chromatographic detection is a vital tool in the evaluation of protein purity. A case in point is High Performance Liquid Chromatography (HPLC), a technique utilized to ascertain protein purity by disentangling the protein from any co-existing impurities and subsequently detecting and quantifying the protein content within the sample.
4. Sedimentation Velocity Methods
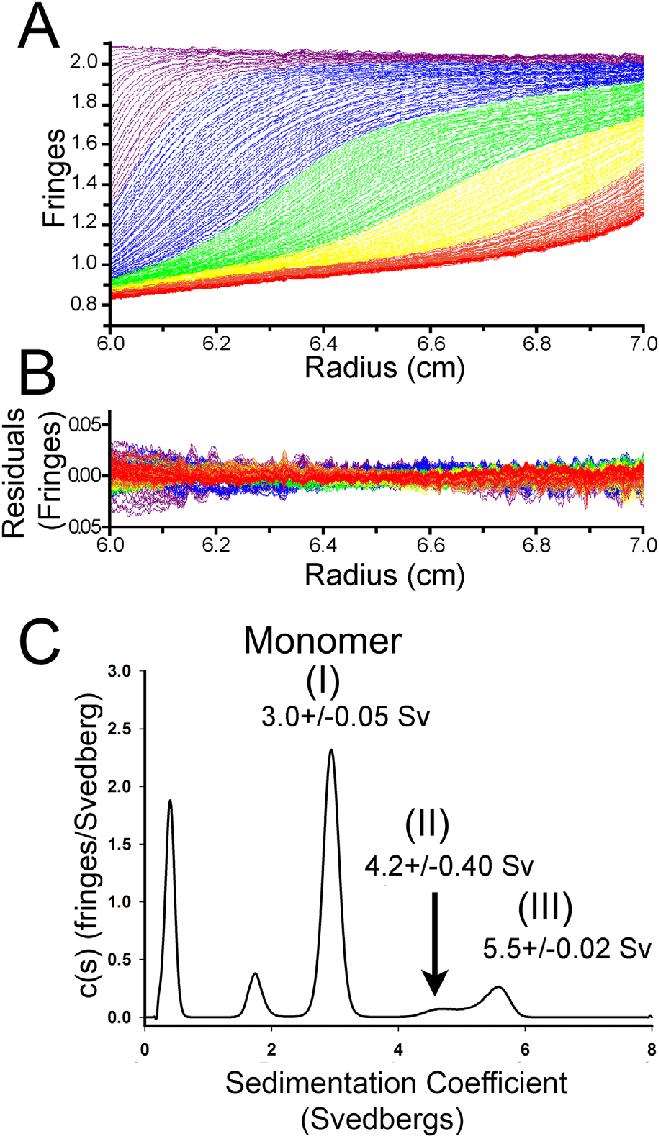 Fig 5. Sedimentation velocity analytical ultracentrifugation analysis of Spa47 oligomerization. (Burgess, J. L., et al.; 2016)
Fig 5. Sedimentation velocity analytical ultracentrifugation analysis of Spa47 oligomerization. (Burgess, J. L., et al.; 2016)
Analytical ultracentrifugation utilizing the sedimentation velocity method is a highly effective technique applied in the examination of macromolecules, significantly proteins. One significant application of this method is in the sphere of protein purity assessments. The fundamental principle is that a truly pure protein sample should exhibit a consistent distribution in terms of size and shape, leading to a unique sedimentation rate. Consequently, the sedimentation velocity method can be a decisive indicator of protein purity by virtue of its ablity to detect this unified sedimentation rate. In contrast, sub-optimal purity in protein samples, due to the existence of extraneous molecular species or contaminants, will produce a broader spectrum of sedimentation rates.
5. Mass Spectrometry Methods
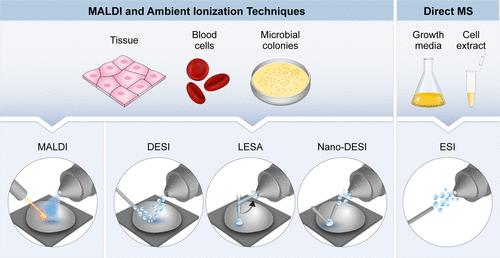 Fig 6. Scheme summarizing the various mass spectrometry ionization techniques for intact protein analysis from crude samples. (Vimer, S., et al.; 2020)
Fig 6. Scheme summarizing the various mass spectrometry ionization techniques for intact protein analysis from crude samples. (Vimer, S., et al.; 2020)
Mass spectrometry is a robust and increasingly utilized analytical technology in the domain of protein purity analysis. This technique allows for the evaluation of the protein sample's purity by scrutinizing the existence of alternate proteins or contaminants. A plethora of mass spectrometry methodologies involves the ionization of the protein specimen, followed by its propelling through a mass analyzer. Owing to its potential for high sensitivity and precision, mass spectrometry is considered indispensable for evaluating protein purity. This is particularly pertinent in the pharmaceutical sector, which necessitates superior levels of protein purity for the manufacture of drug products. The capabilities of mass spectrometry are not inhibited by concentration levels, as it can identify and quantify contaminants, even when present in minuscule concentrations.
6. Light Scattering Methods
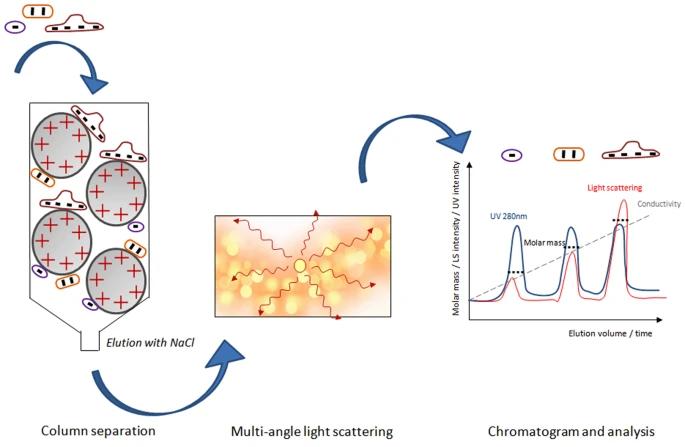 Fig 7. Schematic illustration of the IEX-MALS method. (Amartely, H., et al.; 2018)
Fig 7. Schematic illustration of the IEX-MALS method. (Amartely, H., et al.; 2018)
In characterizing protein purity, light scattering techniques prove inherently valuable in assessing protein samples' purity and homogeneity levels. These techniques operate based on light quantification scattered upon exposure of the protein solution to a light source, typically a laser. Notably, diverse light scattering techniques such as dynamic light scattering (DLS) and static light scattering (SLS) exist, each with distinctive operational mechanisms. For instance, DLS gauges the time-dependent variations in scattered light intensity, offering insights into the size and morphology of protein molecules. Conversely, SLS emphasizes assessing the cumulative intensity of scattered light, thereby delivering critical information regarding protein molecular weight and conformational structure.
Sample Requirement
| Sample Type |
Protein |
Cell |
Animal tissue |
Plant Tissue |
Blood (EDTA added) |
Serum |
Urine |
Microbes |
| Quantity |
100 ug |
2X107 cells |
1g |
200 mg |
1mL |
0.2-0.5 mL |
2 mL |
Dry weighed: 200 mg |
Service Workflow
Want to Learn More?
As a leading specialist in scientific research solutions, Creative Proteomics provides pioneering services in protein purity determination. Leveraging cutting-edge laboratory technology and innovative methods, our expert team of professionals ensures precision and reliability in the results. The primary objective of Creative Proteomics is to expedite our clients' research or analytical objectives. Through our services, our clientele can obtain critical insights into their protein samples' quality that can propel their research and development pursuits, thereby advancing meaningful scientific breakthroughs. Do not hesitate to reach out for further information.
References
- Rhodes, D. G., & Laue, T. M. Chapter 38 Determination of Protein Purity. Guide to Protein Purification, 2nd Edition. 2009, 677–689.
- Conrads, T. P., et al.; Sampling and analytical strategies for biomarker discovery using mass spectrometry. BioTechniques. 2006, 40(6), 799–805.
- Tinoco, A., et al.; Proteins as Hair Styling Agents. Applied Sciences. 2021, 11(9), 4245.
- Calef, S.T., et al.; Purification of Modified Therapeutic Proteins Available on the Market: An Analysis of Chromatography-Based Strategies. Frontiers in Bioengineering and Biotechnology. 2021, Volume 9.
- Burgess, J. L., et al.; Spa47 is an oligomerization-activated type three secretion system (T3SS) ATPase fromShigella flexneri. Protein Science. 2016, 25(5), 1037–1048.
- Vimer, S., et al.; Mass Spectrometry Analysis of Intact Proteins from Crude Samples. Analytical Chemistry. 2020, 92(19), 12741–12749.
- Amartely, H., et al.; Coupling Multi Angle Light Scattering to Ion Exchange chromatography (IEX-MALS) for protein characterization. Scientific Reports. 2018, 8(1).
Related Sections
Services
Applications
For research use only, not intended for any clinical use.

 Fig 1. Determination of Protein Purity. (Rhodes, D. G., & Laue, T. M.; 2009)
Fig 1. Determination of Protein Purity. (Rhodes, D. G., & Laue, T. M.; 2009) Fig 2. Combination of activity-based protein profiling (ABPP) and mass spectrometry (MS)based proteomic profiling to identify specific activities within complex human samples. (Conrads, T. P., et al.; 2006)
Fig 2. Combination of activity-based protein profiling (ABPP) and mass spectrometry (MS)based proteomic profiling to identify specific activities within complex human samples. (Conrads, T. P., et al.; 2006) Fig 3. Protein purity analysis by SDS-PAGE in a 12.5% polyacrylamide gel. (Tinoco, A., et al.; 2021)
Fig 3. Protein purity analysis by SDS-PAGE in a 12.5% polyacrylamide gel. (Tinoco, A., et al.; 2021) Fig 4. General scheme for the purification of commercial PEGylated proteins by chromatography. (Calef, S.T., et al.; 2021)
Fig 4. General scheme for the purification of commercial PEGylated proteins by chromatography. (Calef, S.T., et al.; 2021) Fig 5. Sedimentation velocity analytical ultracentrifugation analysis of Spa47 oligomerization. (Burgess, J. L., et al.; 2016)
Fig 5. Sedimentation velocity analytical ultracentrifugation analysis of Spa47 oligomerization. (Burgess, J. L., et al.; 2016) Fig 6. Scheme summarizing the various mass spectrometry ionization techniques for intact protein analysis from crude samples. (Vimer, S., et al.; 2020)
Fig 6. Scheme summarizing the various mass spectrometry ionization techniques for intact protein analysis from crude samples. (Vimer, S., et al.; 2020) Fig 7. Schematic illustration of the IEX-MALS method. (Amartely, H., et al.; 2018)
Fig 7. Schematic illustration of the IEX-MALS method. (Amartely, H., et al.; 2018)
