Cross-Linking Mass Spectrometry (XL-MS)
 Creative Proteomics are offering cross-linking mass spectrometry (CL-MS), a robust and flexible method for probing PPI binding interfaces and defining protein-protein interactions (PPIs). CL-MS is capable of providing medium-resolution structural information by identifying proximate amino acid pairs (including transient or weak interactions) that are covalently linked by specific lengths of chemical cross-linkers. And the obtained information is further utilized to build direct PPI networks of protein complex as well as mapping PPI binding interfaces.
Creative Proteomics are offering cross-linking mass spectrometry (CL-MS), a robust and flexible method for probing PPI binding interfaces and defining protein-protein interactions (PPIs). CL-MS is capable of providing medium-resolution structural information by identifying proximate amino acid pairs (including transient or weak interactions) that are covalently linked by specific lengths of chemical cross-linkers. And the obtained information is further utilized to build direct PPI networks of protein complex as well as mapping PPI binding interfaces.
How does XL-MS work?
In the XL-MS experiment, the cross-linking reaction is performed at near-native conditions, and chemical reagents and/or UV light are used to form covalent bonds between side chains of amino acids. The artificial bonds have defined lengths and physically bind proximal amino acid residues. Once the cross-linked proteins and protein complexes are formed, enzymatic digestion of the protein/complex is usually performed and the sample is analyzed by high-resolution mass spectrometry (MS). The approach allows the identification of the cross-linked amino acids via database searching, yielding conclusions about their spatial proximity in the protein assembly, involving the sequence assignment of cross-linked peptides as well as localization of specific cross-linked amino acid residues. The physical interaction data derived from XL-MS can be used for structural biology and computational modeling studies. According to the spatial proximity of cross-linked sites, the information of protein binding interfaces, protein conformations, protein dynamics, and novel partnerships can be obtained.
Advantages of the XL-MS method
As a complementary method to support other techniques, chemical XL-MS based on cross-linking reagents provide the unique advantage of capturing both native protein interactions and structures in a physiologically relevant subcellular context.
- Compared to other protein structural techniques, the major benefit of XL-MS is that it allows for a comprehensive snapshot of the protein landscape under minimum interference conditions.
- XL-MS can be completed in a few days, making it an attractive alternative to high-resolution protein structure techniques, such as cryo-electron microscopy (cryo-EM), X-ray crystallography, and native MS.
- The XL-MS experiment requires only a small amount of protein.
Service offering in Creative Proteomics
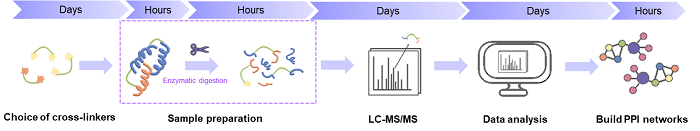
With robust cross-linking reagents, ultra-high performance mass spectrometers, and powerful bioinformatics tools, we utilize a specialized XL-MS approach for defining virus-host PPI networks and protein complexes that are involved in viral pathogenesis. Our services focus on the five key steps of the XL-MS experiment, including choice of cross-linkers, sample preparation, MS analysis, data processing, as well as structural characterization and protein interaction mapping.
Workflow of the XL-MS method
Benefits of our services
- An optimization & scale-up platform
- Professional technical support
- A rapid and tailored solution for specific research applications
- Complete analytical study reports
XL-MS has become one of the most promising MS-based approaches for three-dimensional (3D) structure analysis of large and transient proteins and protein complexes. With the improvements and innovations in each aspect, it has become possible to analyze non-targeted protein complexes and define PPI landscapes at the systems level. Creative Proteomics is a leading provider in the field of viral proteomics services. Based on our XL-MS platform, we can help customers give insight into the progression and spread of a viral infection. For more information on how we can help you, please feel free to contact us. We look forward to providing services for your next project.
References
- Piersimoni, L., & Sinz, A. (2020). "Cross-linking/mass spectrometry at the crossroads." Analytical and Bioanalytical Chemistry, 412, 5981-5987.
- Yu, C., & Huang, L. (2018). "Cross-linking mass spectrometry (XL-MS): An emerging technology for interactomics and structural biology." Analytical chemistry, 90(1), 144.
* For research use only.

 Creative Proteomics are offering cross-linking mass spectrometry (CL-MS), a robust and flexible method for probing PPI binding interfaces and defining protein-protein interactions (PPIs). CL-MS is capable of providing medium-resolution structural information by identifying proximate amino acid pairs (including transient or weak interactions) that are covalently linked by specific lengths of chemical cross-linkers. And the obtained information is further utilized to build direct PPI networks of protein complex as well as mapping PPI binding interfaces.
Creative Proteomics are offering cross-linking mass spectrometry (CL-MS), a robust and flexible method for probing PPI binding interfaces and defining protein-protein interactions (PPIs). CL-MS is capable of providing medium-resolution structural information by identifying proximate amino acid pairs (including transient or weak interactions) that are covalently linked by specific lengths of chemical cross-linkers. And the obtained information is further utilized to build direct PPI networks of protein complex as well as mapping PPI binding interfaces. 