Viral Metagenomic and Bioinformatics Analysis
Since many viruses cannot be cultivated in the laboratory, historic methods have limited the study of viruses in their natural environment, such as cataloging of members and the understanding of the communities' function. Besides, it is a challenge to analyze viral data due to the considerable variability in viruses and the relative scarcity of viral genetic material in samples. There is low coverage of viral diversity in current databases. And it is estimated that a vast majority of virus taxa are yet to be described and classified. Novel DNA sequencing techniques, known as “Next-Generation Sequencing” (NGS) techniques is a high-throughput, impartial tool with numerous attractive features compared to established methods for viral applications in research and diagnostic settings. With the development of NGS, the sequence-independent amplification followed by high-throughput sequencing enables to sequence all viral genomes in a sample without previous knowledge about their nature. Viral metagenomics has benefited greatly from this advanced technology, allowing the discovery of completely new viral species that directly from environmental samples.
 Creative Proteomics' viral metagenomic sequencing service applies whole genome shotgun metagenomic sequencing based on an Illumina HiSeq platform and an assembly-first protocol, which can be used in a broad range of applications. The shotgun metagenomic sequencing has the advantages of high throughput and high coverage, allowing the analysis of genetic material from environmental samples without the prior isolation and cultivation of individual virus. Additionally, we also provide bioinformatic analysis services, including gene predictions, functional annotations, and taxonomic annotations. With the help of our expertise in shotgun sequencing and bioinformatics, our customers can explore the rich genetic repertoire of viral communities and further identify the species, genes, and pathways represented in their samples.
Creative Proteomics' viral metagenomic sequencing service applies whole genome shotgun metagenomic sequencing based on an Illumina HiSeq platform and an assembly-first protocol, which can be used in a broad range of applications. The shotgun metagenomic sequencing has the advantages of high throughput and high coverage, allowing the analysis of genetic material from environmental samples without the prior isolation and cultivation of individual virus. Additionally, we also provide bioinformatic analysis services, including gene predictions, functional annotations, and taxonomic annotations. With the help of our expertise in shotgun sequencing and bioinformatics, our customers can explore the rich genetic repertoire of viral communities and further identify the species, genes, and pathways represented in their samples.
Service Offerings
Creative Proteomics offers one-stop service about viral metagenomic sequencing, including the sample pre-processing, bacteriophage isolation, viral nucleic acid preparation to the result reporting. With highly experienced laboratory professionals and strict quality procedures, each sample at each processing step is obtained enough care to ensure the integrity of customer's results.
Viral metagenomic sequencing
Creative Proteomics' viral metagenomic sequencing service is based on the Illumina HiSeq sequencing system. We can not only process viral nucleic acid, but also accept a variety of types of samples from different environments.
Workflow of viral metagenomic sequencing:
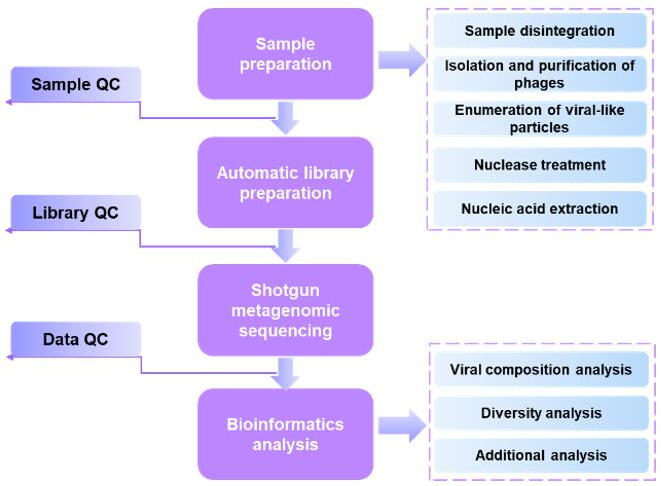
Bioinformatic analysis
Creative Proteomics also offers a range of standard and customized bioinformatics pipelines for our customers' whole genome sequencing projects. Standard bioinformatics pipelines include viral composition analysis, diversity analysis and additional analysis.
Workflow of bioinformatic analysis:
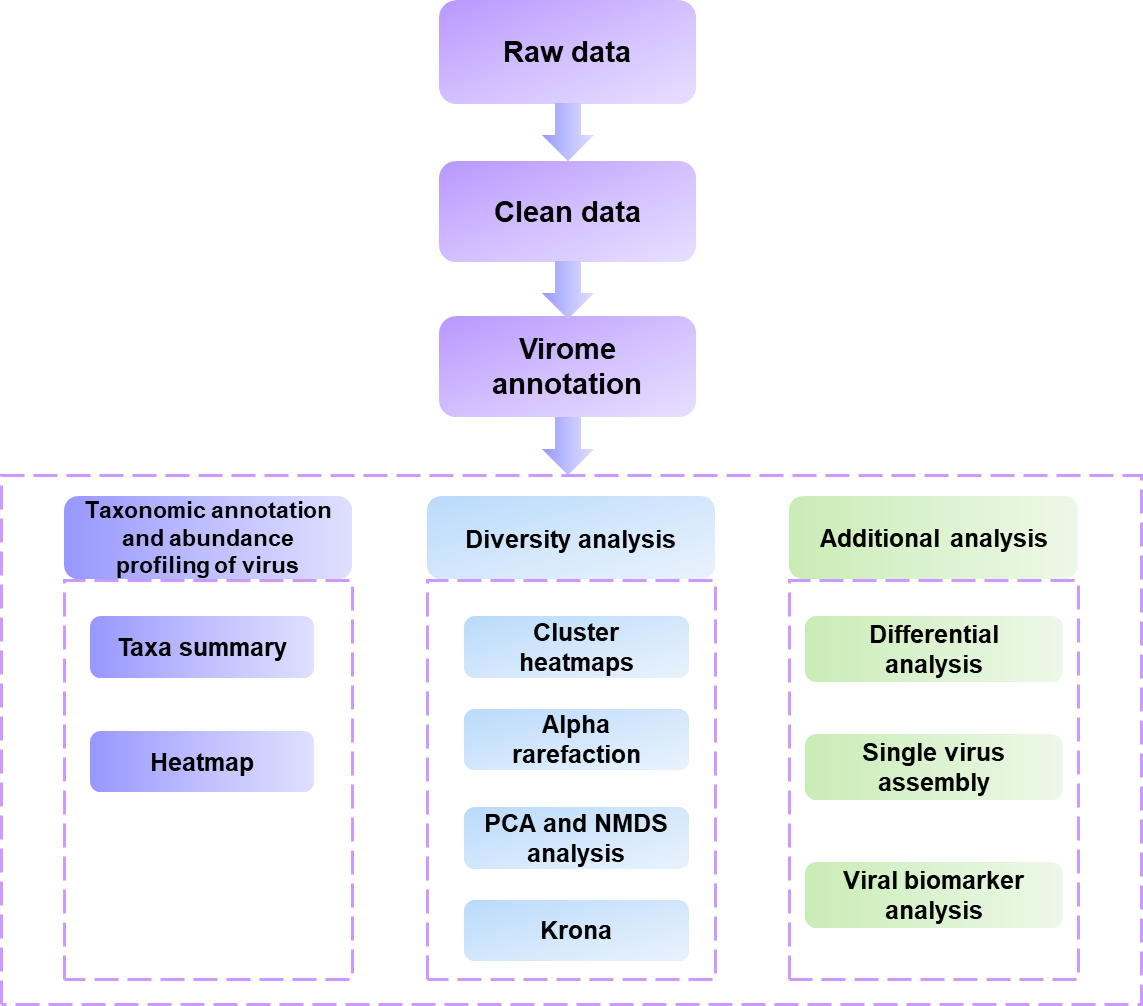
Customized bioinformatics pipelines suit customers' unique projects and you have direct access to our staff and prompt feedback to your inquiries.
Creative Proteomics offers viral metagenomic sequencing service based on an Illumina HiSeq platform. We have a team of highly experienced laboratory professionals in metagenomic sample preparation and sequencing. With efficient standard workflows, we can enhance the generation of data from low-abundance species. More importantly, we provide comprehensive and cost-effective bioinformatics analysis, which is designed according to the specific of our customers' research needs. If you are interested in our services, please contact us for more details.
* For research use only.

 Creative Proteomics' viral metagenomic sequencing service applies whole genome shotgun metagenomic sequencing based on an Illumina HiSeq platform and an assembly-first protocol, which can be used in a broad range of applications. The shotgun metagenomic sequencing has the advantages of high throughput and high coverage, allowing the analysis of genetic material from environmental samples without the prior isolation and cultivation of individual virus. Additionally, we also provide bioinformatic analysis services, including gene predictions, functional annotations, and taxonomic annotations. With the help of our expertise in shotgun sequencing and bioinformatics, our customers can explore the rich genetic repertoire of viral communities and further identify the species, genes, and pathways represented in their samples.
Creative Proteomics' viral metagenomic sequencing service applies whole genome shotgun metagenomic sequencing based on an Illumina HiSeq platform and an assembly-first protocol, which can be used in a broad range of applications. The shotgun metagenomic sequencing has the advantages of high throughput and high coverage, allowing the analysis of genetic material from environmental samples without the prior isolation and cultivation of individual virus. Additionally, we also provide bioinformatic analysis services, including gene predictions, functional annotations, and taxonomic annotations. With the help of our expertise in shotgun sequencing and bioinformatics, our customers can explore the rich genetic repertoire of viral communities and further identify the species, genes, and pathways represented in their samples.
