Quantitative Proteomic Analysis Services
Creative Proteomics provides global proteome profiling services based on label-free or label-based proteomic analysis methods, including isobaric tags for relative and absolute quantification (iTRAQ)-based proteomic analysis, tandem mass tag (TMT)-based proteomic analysis, stable isotopic labeling with amino acids in cell culture (SILAC)-based proteomic analysis, and label-free quantitative proteomic analysis. Each of these methods has its own advantages. We are committed to providing you with the best strategy according to the specific project needs of our clients.
Overview of labeling method for quantitative proteomics
With respect to sample preparation, there are a series of labeling methods for quantitative proteomics, including label-free or label-based methods. The label-based proteomic analysis provides higher relative quantification accuracy, which allows identifying minute differences between samples or experimental conditions. Additionally, they can also be applied to investigate time-resolved profiling of protein abundances as well as dynamic protein turnover. However, they are only suitable for in vitro or cell-culture-based research, and their sample preparation is laborious and time-consuming as they require specialized cell lines and special growth media. For instance, SILAC-based quantitative proteomics enables highly accurate relative quantification by comparing the intensity ratio between isotope-labeled peptides and unlabeled peptides. In the SILAC-based experiment, a light (such as C12, N14) or a heavy amino acid (such as C13, N15) are needed to add into the culture medium that metabolically incorporates into cellular proteins after several cell generations. Furthermore, TMT labels are often added to the specimen at a late stage of sample preparation. The method has been widely applied to increase clinical settings or throughput, suitable for clinical specimens of different biological origins.
Compared with label-based methods, label-free proteomics is relatively inexpensive and less time-consuming. In the label-free quantitative proteomic analysis, proteins are digested into a peptide mixture using proteases, then the samples are analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The relative protein abundance is determined by chromatographic peak intensity measurements or spectral counting. In recent years, due to its broad adaptability and the development of user-friendly software, label-free proteomics is becoming more and more popular.
Service offering in Creative Proteomics
In general, global proteome profiling workflow includes cellular lysis, protein extraction, protein digestion and separation, and MS/MS analysis. During the process, the labeling strategies for quantitative proteomics can be introduced at multiple time points. Metabolic labeling is usually added into initial cell culture or animal model samples at an early stage of sample preparation. Following cell lysis, the proteins can be spiked in or labeled enzymatically during digestion. Although we haven't provided the services yet, we're working on it. Further downstream, peptides can be labeled isobarically. In addition, the label-free approach requires less pre-processing. And the protein quantitation achieved during or after data analysis.
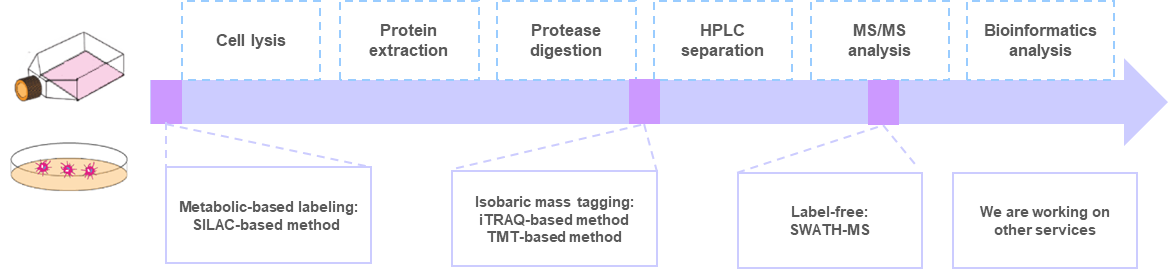 Fig1. Incorporation of labeling method into a typical proteomics workflow.
Fig1. Incorporation of labeling method into a typical proteomics workflow.
Creative Proteomics is an international contract research organization (CRO) providing viral proteomic, metabolomic and other analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. We are dedicated to providing custom experimental design and professional technical support according to customers' specific project needs. If you are interested in our services, please don't hesitate to contact us. We are glad to cooperate with you and witness your success!
Reference
- Simanjuntak, Y., et al. (2021). "Top-Down and Bottom-Up Proteomics Methods to Study RNA Virus Biology." Viruses, 13(4), 668.
* For research use only.

 Fig1. Incorporation of labeling method into a typical proteomics workflow.
Fig1. Incorporation of labeling method into a typical proteomics workflow.