Viral Short-Read RNA Sequencing Service
Creative Proteomics is a good partner of pharmaceutical companies and research institutions. Here, we provide a robust viral transcriptomic analysis using optimized short-read Illumina RNA sequencing (RNA-seq) to provide new insights into the study of viruses.
Short-read sequencing (SRS)
Released in the mid-2000s, the next-generation short-read sequencing (SRS) technology allows the sequencing of millions of DNA fragments simultaneously. SRS has revolutionized genomics and transcriptomics with its high throughput and low cost. The huge amount of reading data generated by SRS has enabled the sequencing of entire genomes of various organisms at an unprecedented rate. In addition to some genome projects, SRS has also been widely applied to study the transcriptome of various organisms, including viruses. At present, SRS RNA-seq offered by Illumina technology is the most widely used and represents the standard for transcriptome profiling. The Illumina technology is characterized by high base accuracy and coverage, providing a great tool to identify transcriptional end sites (TESs), transcriptional start sites (TSSs), splice junctions, and RNA editing. Illumina technology appears to be at the forefront of viral sequencing, being able to interrogate the complex transcriptomes of many viruses.
Service offering in Creative Proteomics
Based on Illumina technology and instruments (including MISeq and HiSeq), we offer optimizing short-read sequencing for viral transcriptomic analysis. Our service is one-stop and customized to meet customers' special requirements. Based on the analysis objectives of customers' projects, we are able to offer two strategies for building the sequencing library, one based on the isolation of polyadenylated RNA from total RNA (polyA (+) RNA) and another based on the removal of highly enriched ribosomal RNA (rRNA-depleted samples). Subsequently, RNA fragments from these two options are used as templates for the synthesis of cDNA, the terminal repair is performed, Illumina-compatible adapters are ligated, and individual samples are indexed for multiplexed sequencing on Illumina platforms. Given that viral transcripts are usually less abundant than those of the host, we combine the improved short-read Illumina RNA-seq workflows with targeted enrichment of viral nucleic acids. As a result, our capability can lead to new biological findings that cannot be identified by conventional approaches.
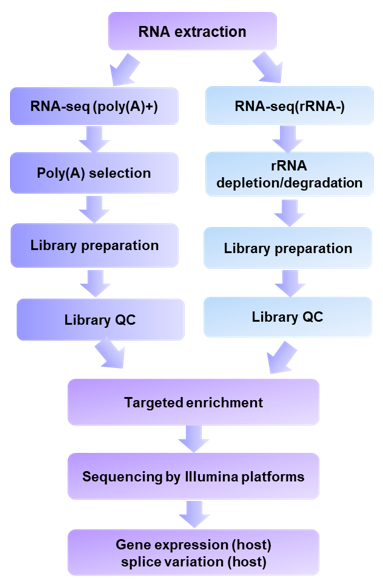 Fig1. Short-read Illumina RNA-seq workflows.
Fig1. Short-read Illumina RNA-seq workflows.
Advantage of our services
- Sequencing the viral transcriptome using optimizing short-read seq
- Rich experience in dealing with viral RNA sequencing
- Provide workflows and analysis tools for RNA sequencing customized for your research
- Strictly keep confidential the client's project information and experimental data
Creative Proteomics' platforms hold great potential for viral transcriptome sequencing. In addition to studying the virus itself, our service can also be applied to study the interaction between virus and the infected host. We put our heart and soul into supporting our global customers with competitive and professional services. Whatever the scale and complexity of your project for a professional, please feel free to contact us for reliable and highly customized solutions.
Reference
- Depledge, D. P., et al. (2018). "Going the distance: optimizing RNA-Seq strategies for transcriptomic analysis of complex viral genomes." Journal of virology, 93(1), e01342-18.
* For research use only.

 Fig1. Short-read Illumina RNA-seq workflows.
Fig1. Short-read Illumina RNA-seq workflows.