Viral SILAC-Based Proteomic Service
Stable isotopic labeling with amino acids in cell culture (SILAC) is an MS-based approach for quantitative proteomics that relies on metabolic labeling of the whole cellular proteome. After labeled with "light" or "heavy" forms of amino acids, the proteomes of different cells grown in cell culture are differentiated through MS. Due to the advantages of in vivo labeling, SILAC labeling better retains various biological characteristics of the protein, and the labeling effect is not biased. Therefore, SILAC is well-suited for a wide range of biological applications, including cancer, embryonic stem cells, and virus-host protein interactions.
How does SILAC-based quantitative proteomics work?
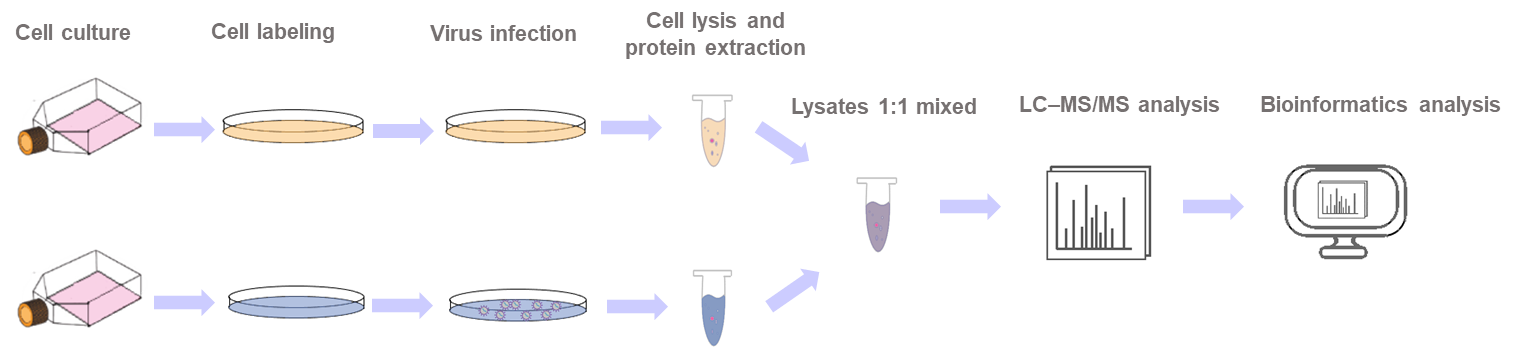
SILAC is considered one of the most accurate quantitative approaches using cell culture systems to study the dynamics of biological processes. In the SILAC-based experiment, the cells are fed with culture mediums containing a light (such as C12, N14) or a heavy amino acid (such as C13, N15). After several generations, proteins in the cells can be labeled by the respective light and heavy amino acid. Then protein lysates are mixed at equimolar ratios prior to being analyzed by mass spectrometry (MS). The relative quantification of the peptides is accomplished by comparing the area of isotopic peaks in the primary MS, and the peptides are sequenced by a secondary spectrogram to identify the proteins.
Advantages of SILAC-based proteomic analysis
- High sensitivity and small sample size requirements.
- High labeling rate with reproducible and reliable results.
- Hundreds to thousands of proteins can be identified and quantified simultaneously.
- As an in vivo labeling technology, SILAC is highly active and can reveal the state of the sample more truly.
SILAC-based proteomic analysis of virus infection
SILAC-based quantitative proteomic approaches have been widely applied to profile global translation rates of viral and host proteins for multiple (+)RNA viruses, including HIV, DENV, and hepatitis C virus (HCV). Early in 2010, SILAC coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS) had been applied to map the nucleolar protein profiles of coronavirus-infected bronchitis virus. Among the 378 identified cellular proteins, one viral protein ( N protein) was found to be localized in the nucleolus and firstly reported. Furthermore, the quantitative proteomic method has been employed to study influenza A virus–nucleolar interactions in human T cells, and a series of virus-encoded proteins were identified, including nucleolar proteins. A study used SILAC combined with 2D gel electrophoresis and MALDI-TOF-MS to investigate the virus-host interactions of pseudorabies virus (PrV) infected bovine cells. By comparing the infected cells and non-infected cells, a total of 55 proteins with significantly altered levels were identified.
Service offering
Based on excellent experts, well-established proteome technology platforms, and professional bioinformatics analysis, we are able to provide our customers with a one-stop service in SILAC quantitative proteomics. Combined with other technologies, such as co-immunoprecipitation, quantitative immunoprecipitation combined with knockdown (QUICK), and RNA interference (RNAi), we can quantitatively screen and identify the virus-host protein interactions. Generally, our comprehensive services include cell culture, virus infection, cell labeling, protein extraction and digestion, LC-MS/MS analysis, and bioinformatics analysis.
Workflow of SILAC-based proteomic analysis of virus infection
Creative Proteomics is a forward-looking research institute as well as a leading custom service provider in the field of viral proteomics. We provide custom experimental design and professional technical support according to customers' specific project needs. If you are interested in our services, please don't hesitate to contact us. We are glad to cooperate with you and to witness your success!
Reference
- Simanjuntak, Y., et al. (2021). "Top-Down and Bottom-Up Proteomics Methods to Study RNA Virus Biology." Viruses, 13(4), 668.
* For research use only.

