Proteomics in DENV Research
As the most widespread mosquito-borne viral disease, dengue virus (DENV) infection is a worldwide public health threat, especially in developing countries. Due to the lack of broadly effective vaccines as well as antiviral drugs, the rise in dengue virus infection rates is worrying. Besides, the knowledge about the pathogenesis and progression of DENV infection has remained elusive. Creative Proteomics is an international contract research organization (CRO) providing viral proteomic, metabolomic and other analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. We are dedicated to providing a series of proteomic techniques to help customers quickly and comprehensively investigate viral proteomics. Here, we will briefly review proteomics studies of DENV in recent years.
Dengue virus (DENV)
Dengue virus (DENV) is an enveloped, single-stranded positive-sense RNA virus belonging to the genus Flavivirus in the family Flaviviridae. Similar to the Zika virus (ZIKV), DENV is transmitted to humans primarily by the Aedes genus mosquito. The virus can lead to a serious health problem called dengue fever, with a range of symptoms ranging from mild to severe. DENV can encode ten proteins, including three structural proteins (C, prM and E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5). Each viral protein has a specific function and facilitates the production of new virus particles, and further affects cellular events.
Proteomic study of dengue virus (DENV)
To explore the pathogenesis of DENV infection and the interaction between the virus and host cells, proteomic analysis is a powerful method. To date, current proteomic analysis for studying DENV has focused on proteome changes after viral infection, the interaction between non-structural viral proteins and host cell proteins as well as diagnostics.
Quantitative proteomics applied to evaluate host cell responses to DENV-2 infection
Currently, DENV can be divided into four distinct and closely related serotypes (share approximately 65% of their genomes), including DENV-1, DENV-2, DENV-3, DENV-4. Since DENV-2 is the most widespread of all DENV serotypes, researchers applied quantitative proteomics to evaluate the protein changes and modifications in the K562 cell line during DENV-2 infection. The study revealed important virus-induced changes that cover a wide range of host cellular compartments and processes by analyzing the differentially expressed proteins in the infected cells and the simulated control cells. Furthermore, there are more dramatic changes in protein phosphorylation compared to protein expression.
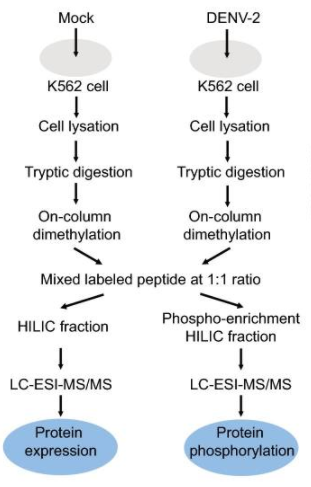 Fig1. Proteomics and phosphoproteomics profiling of DENV-2 infection in human K562 cells. (Han, L., et al, 2019).
Fig1. Proteomics and phosphoproteomics profiling of DENV-2 infection in human K562 cells. (Han, L., et al, 2019).
Quantitative comparative proteomics reveal biomarkers for dengue disease severity
DENV can lead to a serious health problem called dengue fever. According to the World Health Organization (WHO), the disease severity can be graded as dengue fever (DF) and dengue hemorrhagic fever (DHF) grades 1–4, depending on its clinical manifestations. DHF has a higher mortality rate than DF. It's critical to identify different molecular biomarkers to predict DHF patients from DF since they have the same pathogen and clinical characteristics. Researchers applied a quantification (TMT)-based quantitative proteomics approach to identify differentially expressed proteins between DF and DHF patients. Among 441 altered proteins, PLAT, LAMB2, F9, VCAM1, FGL1, MFAP4, and GLUL could be potential molecular biomarkers of predicting DHF from DF patients.
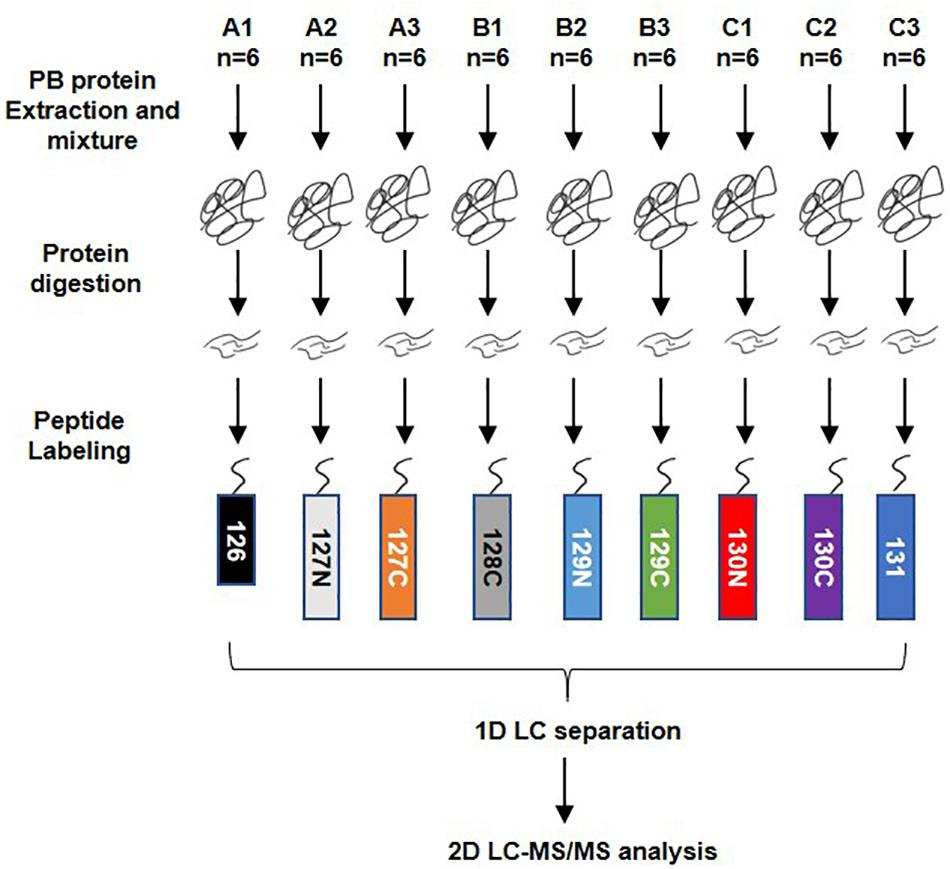 Fig2. Workflow of TMT quantitatively proteomics analysis. A total of 54 serum samples from dengue haemorrhagic fever (DHF), Dengue fever (DF), and healthy groups were collected, numbered A, B, and C, with 18 in each group.( Miao, M., et al, 2019)
Fig2. Workflow of TMT quantitatively proteomics analysis. A total of 54 serum samples from dengue haemorrhagic fever (DHF), Dengue fever (DF), and healthy groups were collected, numbered A, B, and C, with 18 in each group.( Miao, M., et al, 2019)
Based on professional scientists and advanced platforms, Creative Proteomics is capable of providing a series of proteomic techniques to help customers quickly and comprehensively investigate viral proteomics, including viral protein structure detection, global proteome profiling, mass spectrometry-based viral proteomics, and so on. For more information on how we can help you, please feel free to contact us or directly send us an inquiry.
References
- Han, L., et al. (2019). "Quantitative Comparative Proteomics Reveal Biomarkers for Dengue Disease Severity." Frontiers in microbiology, 10, 2836.
- Miao, M., et al. (2019). "Proteomics profiling of host cell response via protein expression and phosphorylation upon dengue virus infection." Virologica Sinica, 34(5), 549-562.
Related services
* For research use only.

 Fig1. Proteomics and phosphoproteomics profiling of DENV-2 infection in human K562 cells. (Han, L., et al, 2019).
Fig1. Proteomics and phosphoproteomics profiling of DENV-2 infection in human K562 cells. (Han, L., et al, 2019).  Fig2. Workflow of TMT quantitatively proteomics analysis. A total of 54 serum samples from dengue haemorrhagic fever (DHF), Dengue fever (DF), and healthy groups were collected, numbered A, B, and C, with 18 in each group.( Miao, M., et al, 2019)
Fig2. Workflow of TMT quantitatively proteomics analysis. A total of 54 serum samples from dengue haemorrhagic fever (DHF), Dengue fever (DF), and healthy groups were collected, numbered A, B, and C, with 18 in each group.( Miao, M., et al, 2019)