Proteomics in Influenza Virus Research
As one of the common respiratory pathogens, influenza viruses continue evolving and can cause substantial morbidity and mortality, posing an annual threat to public health and animal livestock. Therefore, it is essential to elucidate its pathogenesis and find new treatment methods. In recent years, proteomics has made important advances in the pathogenesis of influenza viruses, involving the dynamic interactions between host cells and influenza viruses, and the post-translational regulation of viral infection. Now, let us take a look at proteomics in the recent study of influenza viruses.
Influenza virus
 Influenza viruses with single-stranded, negative-sense, and segmental RNA genomes, are members of the Orthomyxoviridae family. Influenza viruses are characterized by rapid evolution and can cause respiratory infections in mammals (e.g., humans, pigs, horses) and birds (e.g., chickens, waterfowl). The viruses can be divided into A, B, and C groups according to group-specific antigens. Among them, influenza A viruses (IAVs) belonging to the genus Alphainfluenzavirus, are of zoonotic importance. Based on the proteins on the surface of the virus (including haemagglutinin, H1-H18, and neuraminidase, N1-H11), IAVs can be divided into a series of subtypes. IAVs cause seasonal epidemics and sporadically pandemic outbreaks, posing an economic burden and health threat to humans.
Influenza viruses with single-stranded, negative-sense, and segmental RNA genomes, are members of the Orthomyxoviridae family. Influenza viruses are characterized by rapid evolution and can cause respiratory infections in mammals (e.g., humans, pigs, horses) and birds (e.g., chickens, waterfowl). The viruses can be divided into A, B, and C groups according to group-specific antigens. Among them, influenza A viruses (IAVs) belonging to the genus Alphainfluenzavirus, are of zoonotic importance. Based on the proteins on the surface of the virus (including haemagglutinin, H1-H18, and neuraminidase, N1-H11), IAVs can be divided into a series of subtypes. IAVs cause seasonal epidemics and sporadically pandemic outbreaks, posing an economic burden and health threat to humans.
Recent proteomic study in Influenza virus infection
Character dynamic interactions between host cells and influenza viruses
To investigate dynamic alternations in global protein profiles upon infection of various cell types (such as macrophage, A549 cells, and MDCK cells) with IAVs, a series of proteomic approaches have been used, including two-dimensional electrophoresis MS/MS, SILAC, and iTRAQ. A recent study applied iTRAQ-based proteomic and bioinformatic to quantify human mast cells (HMCs) proteins that may be regulated by differences in the subtypes of IAV virus infection and insight into the relative functional activities of these various proteins. In addition, since posttranslational modifications (PTMs) can affect the virulence of the virus as well as the host-response, the related research has significance in the viral-host interactions. Proteomics has made important advances in describing the PTMs of influenza viruses, mainly IAVs and their hosts. A recent paper gave a detailed review of the PTMs identified in influenza by using multiple proteomics methodologies, involving phosphorylation, glycosylation, methylation, acetylation, ubiquitination and ubiquitin-like modification, and some types of acylation.
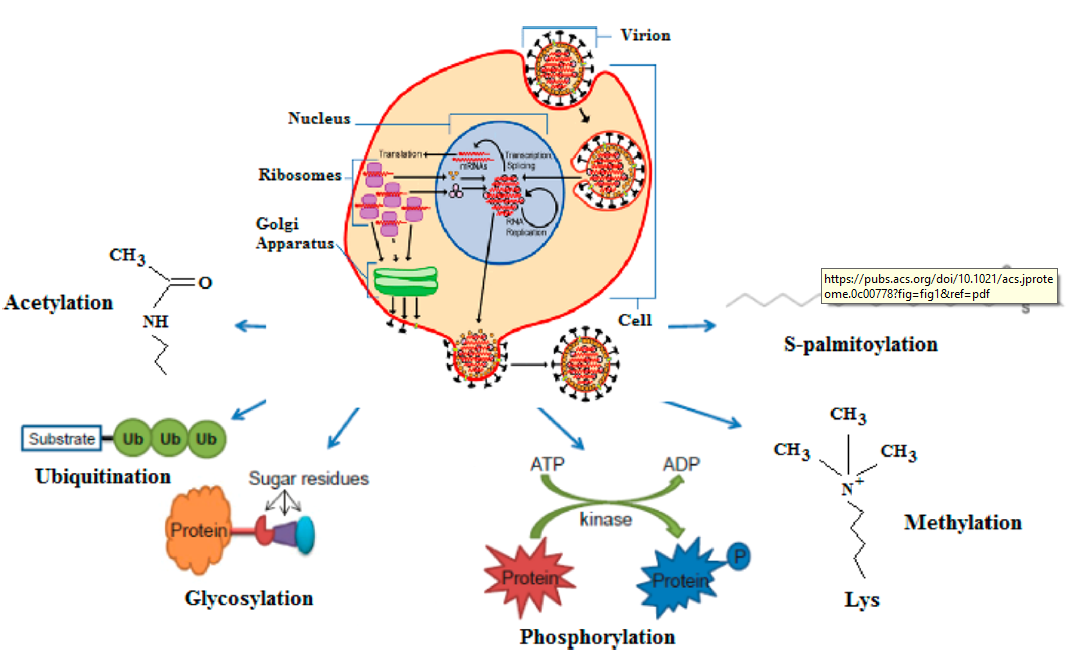 Summary of the PTMs in the interactions between influenza virus and its host from published documents. (Zhang, J.,et al, 2020)
Summary of the PTMs in the interactions between influenza virus and its host from published documents. (Zhang, J.,et al, 2020)
Investigate interspecies reaction and how the virus adapts to humans from other species
Based on metabolic pulse labeling and quantitative mass spectrometry (MS), researchers compared proteome dynamics upon infection of human cells with a bird-adapted IAV and a human-adapted strain. The results showed that the host proteins behaved similarly and the viral protein synthesis was strikingly different, especially for the matrix protein M1. Finally, researchers identified M segment RNA splicing as a host range determinant according to In silico and biochemical evidence. In addition, proteomics technologies were also applied in studies to validate influenza vaccines.
Creative Proteomics is a forward-looking research institute as well as a leading custom service provider in the field of viral proteomics. We are dedicated to providing a series of proteomic techniques to help customers quickly and comprehensively investigate viral proteomics. For more information on how we can help you, please feel free to contact us or directly send us an inquiry.
References
- Katsarou, E. I., et al. (2021). "Applied Proteomics in 'One Health'." Proteomes, 9(3), 31.
- Zhang, J.,et al. (2020). "Proteomics in influenza research: The emerging role of posttranslational modifications." Journal of Proteome Research, 20(1), 110-121.
Related services
* For research use only.

 Influenza viruses with single-stranded, negative-sense, and segmental RNA genomes, are members of the Orthomyxoviridae family. Influenza viruses are characterized by rapid evolution and can cause respiratory infections in mammals (e.g., humans, pigs, horses) and birds (e.g., chickens, waterfowl). The viruses can be divided into A, B, and C groups according to group-specific antigens. Among them, influenza A viruses (IAVs) belonging to the genus Alphainfluenzavirus, are of zoonotic importance. Based on the proteins on the surface of the virus (including haemagglutinin, H1-H18, and neuraminidase, N1-H11), IAVs can be divided into a series of subtypes. IAVs cause seasonal epidemics and sporadically pandemic outbreaks, posing an economic burden and health threat to humans.
Influenza viruses with single-stranded, negative-sense, and segmental RNA genomes, are members of the Orthomyxoviridae family. Influenza viruses are characterized by rapid evolution and can cause respiratory infections in mammals (e.g., humans, pigs, horses) and birds (e.g., chickens, waterfowl). The viruses can be divided into A, B, and C groups according to group-specific antigens. Among them, influenza A viruses (IAVs) belonging to the genus Alphainfluenzavirus, are of zoonotic importance. Based on the proteins on the surface of the virus (including haemagglutinin, H1-H18, and neuraminidase, N1-H11), IAVs can be divided into a series of subtypes. IAVs cause seasonal epidemics and sporadically pandemic outbreaks, posing an economic burden and health threat to humans.  Summary of the PTMs in the interactions between influenza virus and its host from published documents. (Zhang, J.,et al, 2020)
Summary of the PTMs in the interactions between influenza virus and its host from published documents. (Zhang, J.,et al, 2020)