Multi-Omics in EBOV Infection
Ebola virus (EBOV), which causes a fatal haemorrhagic fever syndrome in humans and non-human primates, is one of the world's most dangerous viruses. Ebola virus disease (EVD) caused by EBOV is marked by high levels of virus replication and dissemination, dysregulated immune responses, extensive tissue damage and organ dysfunction as well as disordered coagulation. In order to investigate the complex pathogenesis of EVD, multi-omics approaches have been used to analyze plasma and peripheral blood mononuclear cells (PBMCs) from patients infected with EVD. Compared with individual platforms, multi-omics approaches provide a more complete context for understanding host-response contributions to disease pathology.
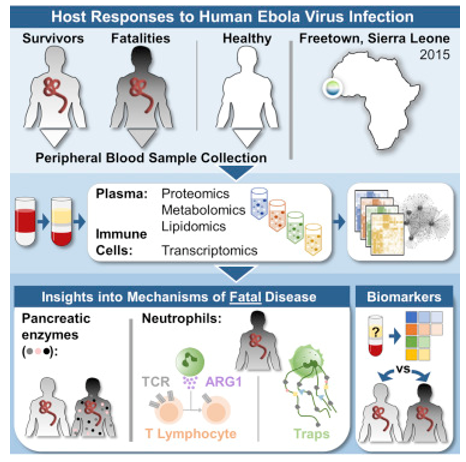 Fig1. Multi-platform' omics analysis of human ebola virus disease pathogenesis. (Eisfeld, A. J., et al, 2017)
Fig1. Multi-platform' omics analysis of human ebola virus disease pathogenesis. (Eisfeld, A. J., et al, 2017)
Multi-omics in EVD
Plasma metabolomics analysis in EVD
According to the plasma metabolome, EVD fatalities were characterized by several major metabolic features, including acute reductions in plasma free amino acids (PFAAs), disrupted sucrose catabolism as well as increased plasma inositol and 2-hydroxybutyric acid. Moreover, plasma lipidomics analyses revealed alterations in lipid subclasses that define fatal and non-fatal EVD. Compared with survivors, there was increased diacylglycerophosphoglycerol, a subset of monoacylglycerophosphoserine (PS), and ceramide species in EVD fatalities. And in fatalities, monoacylglycerolipid and cholesterol ester were significantly increased and decreased.
Plasma proteomics analysis in EVD
Using pathway enrichment analysis, researchers investigated plasma proteomics of EVD. The "Cell Adhesion Molecules" pathway (KEGG hsa04514) was a common feature of EBOV infection as it was enriched in EVD patients compared with healthy controls. There were upregulated components, including adhesion molecules (FCGR3B, FCGR3A, CD14, CD163) and multiple class I major histocompatibility complex antigens, suggesting macrophages and neutrophils were strongly activated in the host response to EVD, especially in fatalities. In addition, the "Pancreatic Secretion" pathway (KEGG hsa04972) was another significantly enriched pathway when compared EVD survivors with fatalities. Increased levels of pancreatic trypsins, regenerating family proteins REG1A and REG3A, and other factors secreted by pancreatic acinar cells were found in the plasma of EVD fatalities.
PBMC RNA-Seq analysis in EVD
Multiscale Embedded Gene Co-expression Network Analysis (MEGENA) can identify modules of co-expressed genes from large transcriptional datasets. Using this method, researchers identified 27 modules that comprise the EVD co-expression network, involving differentially expressed transcripts and post-infection invariant transcripts. The transcriptomics study of PBMCs revealed that EBOV infection strongly induced the expression of antiviral and anti-apoptotic genes, and neutrophils might play an important role in EBOV pathogenicity. Finally, these datasets provided by multi-platform omics analysis are a valuable resource to conduct for the discovery and evaluation of EVD biomarkers and will facilitate the development of diagnostic analyses for EVD.
Through the multi-platform omics analysis of peripheral blood of EVD patients, researchers not only have a deeper understanding of the pathogenesis of EVD, but also provide an effective way for the identification of biomarkers, which is conducive to the further development of prevention or treatment strategies. Creative Proteomics is able to provide transcriptomics, proteomics, and metabolomics experiments and multi-omics joint analysis services. Based on well-established longitudinal cohorts and multi-omics technologies, we can greatly promote the study of many viruses. If you have any needs in this area, please feel free to contact us. We will serve you wholeheartedly.
Reference
- Eisfeld, A. J., et al. (2017). "Multi-platform' omics analysis of human Ebola virus disease pathogenesis." Cell host & microbe, 22(6), 817-829.
Related services
* For research use only.

 Fig1. Multi-platform' omics analysis of human ebola virus disease pathogenesis. (Eisfeld, A. J., et al, 2017)
Fig1. Multi-platform' omics analysis of human ebola virus disease pathogenesis. (Eisfeld, A. J., et al, 2017)