Host Cell scRNA Sequencing Service
To facilitate the identification of host cells containing viral RNA (vRNA) and characterize their transcriptional response, we provide a single-cell RNA sequencing (scRNA-seq) service. Our platform is capable of characterizing transcriptome dynamics in cultured cells infected with a variety of viruses. Through simultaneously quantifying host-cell mRNA and viral RNAabundance within a single cell, proviral and antiviral factors associated with intracellular viral abundance can be identified. Creative Proteomics is a good partner of pharmaceutical companies and research institutions. Our experienced team can provide genomics, transcriptomics, proteomics, metabolomics, and lipidomics experiments as well as in-depth multi-omics data analysis services, greatly accelerating global customers' project progress.
Single-cell RNA-sequencing (scRNA-seq)
scRNA-seq is a technique to study the entire transcriptome at the single-cell level. The principle of scRNA-seq is to perform high-throughput sequencing after efficient amplification of trace transcriptome mRNA in a single cell isolated from multicellular organisms. This technique can effectively solve the problem of cell heterogeneity in tissue samples as well as the problem of transcriptome heterogeneity in cell populations that are covered by conventional RNA-Seq, thus contributing to the discovery of new rare cell types and in-depth understanding of the expression regulation mechanism during development. Despite there are still some limitations, the applications of single-cell RNA-seq, such as identification of host cells containing viral RNA (vRNA) and characterizing their transcriptional response, have promoted a deeper understanding of the genomic diversity of viruses as well as the biology of virus-host interactions.
Service offering in Creative Proteomics
Our service is scalable, based on a single-cell RNA-seq method combined with surface markers for fluorescence-activated cell sorting (FACS). We can simultaneously quantify host-cell mRNA and viral RNA abundance within a single cell. Single-cell RNA-seq requires first isolation and acquisition of the entire transcriptome within a single cell. In order to ensure a sufficient number of high-quality cells from many cell types, we increase the throughput and PCR preamplification. In addition, single-cell isolation is a key step in single-cell RNA-seq, and this step in our service is mainly achieved by FACS. Based on an optimized approach with higher sensitivity and sample selection, the number of viral reads can be maximized. Our service could advance the understanding of the pathogenesis of any viral infection, map an atlas of infected cells, and further promote the investigation of virus-host interactions.
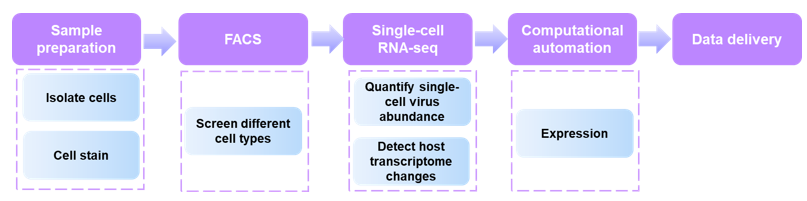 Fig1. FACS-assisted single-cell RNA-Seq workflow on cells from virus-infected and healthy control subjects.
Fig1. FACS-assisted single-cell RNA-Seq workflow on cells from virus-infected and healthy control subjects.
The simultaneous interrogation of host and viral transcriptomes at the single-cell level offers a powerful new tool for the understanding of the biology of virus-host interactions. Creative Proteomics' success lies in constantly optimizing our services. We put our heart and soul into supporting our global customers with competitive and professional services. Whatever the scale and complexity of your project for a professional, please feel free to contact us for reliable and highly customized solutions.
References
- An, P., et al. (2021). "Single-Cell Transcriptomics Reveals a Heterogeneous Cellular Response to BK Virus Infection." Journal of Virology, 95(6), e02237-20.
- Zanini, F., et al. (2018). "Virus-inclusive single-cell RNA sequencing reveals the molecular signature of progression to severe dengue." Proceedings of the National Academy of Sciences, 115(52), E12363-E12369.
* For research use only.

 Fig1. FACS-assisted single-cell RNA-Seq workflow on cells from virus-infected and healthy control subjects.
Fig1. FACS-assisted single-cell RNA-Seq workflow on cells from virus-infected and healthy control subjects.