Viral Transcriptome Sequencing Services
Viruses have small, compact genomes. Detection of the viral transcriptome or viral RNA genome is limited by the low relative abundance of viral RNA relative to the host-derived transcriptome in any given sample. Creative Proteomics is a good partner of pharmaceutical companies and research institutions. Our experienced and professional teams provide robust viral transcriptome sequencing services using a wide range of sequencing technologies, including optimizing short (Illumina) and/or long-read sequencing (PacBio and ONT nanopore). In addition, we also offer a single-cell RNA sequencing (scRNA-seq) service to facilitate the identification of host cells containing viral RNA (vRNA) and characterize their transcriptional response.
The next-generation short-read sequencing (SRS) technology was released in the mid-2000s. SRS has revolutionized genomics and transcriptomics by sequencing millions of DNA fragments simultaneously. At present, SRS RNA sequencing offered by Illumina technology is the most widely used and represents the standard for transcriptome profiling. The third-generation long-read sequencing (LRS) technology emerged in 2011. Currently, there are two widely used LRS technologies, including the PacBio and ONT nanopore platforms. The PacBio and ONT sequencing have the capability to read full-length transcripts and allow the direct investigation of base modifications on both DNA and RNA molecules. Since viruses have small, compact genomes, these organisms are ideal subjects for transcriptome analysis with LRS techniques.
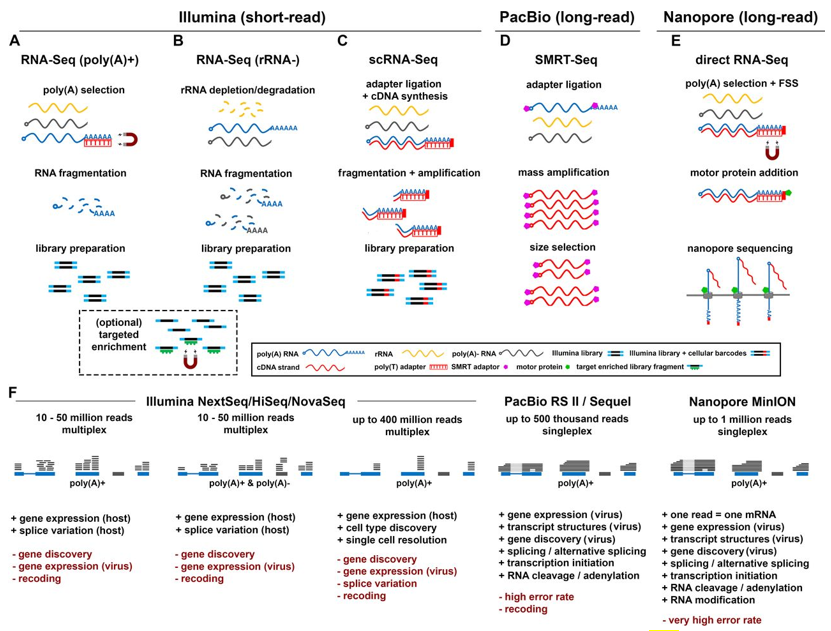 Fig1. Comparison of major RNA sequencing methodologies. (Depledge, D. P., et al, 2018)
Fig1. Comparison of major RNA sequencing methodologies. (Depledge, D. P., et al, 2018)
Viral transcriptome sequencing services in Creative Proteomics
Creative Proteomics offers robust technologies to bring new insights into the study of viral transcriptomes.
When profiling viral transcriptomes by using these sequencing technologies, we understand the exact nature of the information provided by different approaches and pay much attention to four key points.
- The primary focus of the projects, viral and/or host transcripts
- The choice of viral strain and host model
- The goal of the projects, such as to define the transcript isoform landscape and to perform gene expression profiling
- The specific technical and analytical requirements for viral transcriptome studies
Applications of our services
- Identification of novel viral transcripts
LRS can discover the new transcripts, including embedded messenger RNAs (emRNAs), overlapping transcripts, polygenic transcripts, and RNA isoforms (such as splice, TES, and TSS variants). In addition, the technologies can also reveal noncoding RNAs (ncRNAs, such as iRNAs, asRNAs, and encRNAs) and special classes of transcripts (such as near-replication-origin RNAs and nro-like transcripts).
- Elucidation of a novel level of genetic regulation
Transcripts can overlap with each other in a convergent, parallel, or divergent manner. LRS can help to elucidate the role of transcriptional overlaps as well as replication-associated RNA (raRNAs) overlap.
- Exploration of translationally active short uORFs in viral genomes
There are many translationally active short uORFs in viral genomes. Some of these short peptides may be functional, and LRS can help us to explore their specific functions.
- Detection of RNA editing and modification
There are several types of RNA modifications described in the function of the viral transcriptome, including mRNA maturation and evasion of the host's immune response. With the ability to sequence full-length native RNA molecules, the problem of classifying modified nucleotides detected in the overlapping transcripts has been simplified.
If you have questions concerning our services or you'd like to have some more information, please feel free to contact us. We'll get back to you in time.
References
- Tombácz, D., et al. (2020). "Long-read Assays Shed New Light on the Transcriptome Complexity of a Viral Pathogen and on Virus-Host Interaction." bioRxiv.
- Depledge, D. P., et al. (2018). "Going the distance: optimizing RNA-Seq strategies for transcriptomic analysis of complex viral genomes." Journal of virology, 93(1), e01342-18.
* For research use only.

 Fig1. Comparison of major RNA sequencing methodologies. (Depledge, D. P., et al, 2018)
Fig1. Comparison of major RNA sequencing methodologies. (Depledge, D. P., et al, 2018)