mNGS for Virus Detection and Identification
At present, Metagenomic next-generation sequencing (mNGS), as one of the two common methods for virus detection and identification, the detection principle of which is different from real-time fluorescent quantitative PCR(RT-PCR). mNGS not only has incomparable advantages over other methods that enable rapid identification of emerging pathogens, but also provides the basis for accurate diagnosis of other pathogens and mixed infections. Creative Proteomics is dedicated to providing the very best and most progressive service for our worldwide customers based on highly experienced laboratory professionals and efficient standard workflows. Here in Creative Proteomics, besides RT-PCR, we also provide mNGS for virus detection and identification. mNGS is based on the next-generation sequencing technology for unbiased culture-independent pathogen detection. With this technology, our customers have access to detect multiple viruses at the same time as well as discover novel and highly divergent viruses. In addition, if necessary, we can provide the gold-standard in conducting studies of the pathogenicity of viruses for further validation of the sequencing result of customer' viral specimen, such as plaque assay or TCID50 assay.
mNGS for virus detection and identification
Diverse viral communities have crucial roles in the environment and in human health. With the breakout of COVID-19, people get a deeper awareness of the importance of viral research. mNGS is a high-throughput, impartial technology with numerous attractive features compared to historic diagnostic methods, which is widely used in many possible applications in viral research and diagnostic settings. The key advantages include high throughput and high coverage, allowing the analysis of genetic material from environmental samples without the prior isolation and cultivation of individual viruses. This combination of techniques, known as viral metagenomics, improves our understanding of viral diversity, specifically in the detection of novel viruses that cannot be cultured. Because of the decreasing costs of this technique, the application of it in the field of viral research is exponentially increasing.
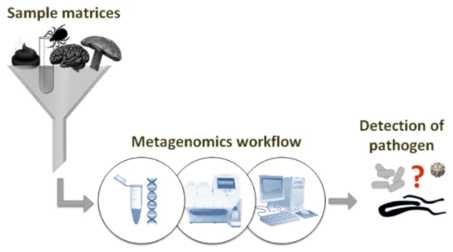 Sketch of the basic idea of a "one serves all" analytical framework. (Wylezich, C., et al., 2018).
Sketch of the basic idea of a "one serves all" analytical framework. (Wylezich, C., et al., 2018).
The advantages of mNGS for viral detection and identification
- Full and rapid identification of viral samples
- Sequence in-depth analysis
- No requirement of the prior isolation and cultivation of individual viruses
- Comprehensive identification of other viruses and mixed infections
The challenges of mNGS for viral detection and identification
- Method standardization
- High platform requirement
- Skilled technicians and bioinformatics analysts are required
- Data storage, protection, analysis, and interpretation
Service offering
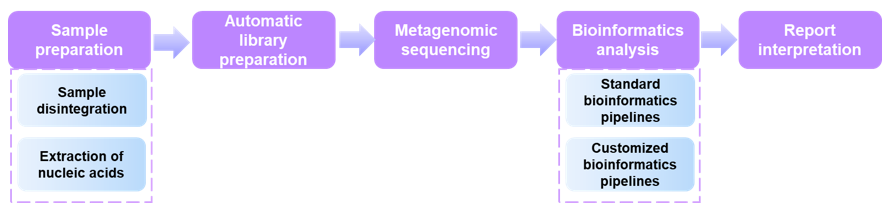
In Creative Proteomics, we provide a one-stop service of mNGS-based virus detection and identification, including sample preparation, automatic library preparation, metagenomic sequencing, bioinformatics analysis, and report interpretation. For unbiased metagenomic sequencing, the sample preparation is a critical part, and is generally comprised of virus enrichment, extraction of nucleic acids, and unbiased amplification. According to different sample types, we offer different strategies, involving the adapted sample treatment and virus enrichment methods. Based on the Illumina HiSeq sequencing system, highly experienced technicians, and efficient standard workflows, we can enhance the generation of data from low-abundance species. More importantly, we have a team of excellent bioinformatics analysts. Therefore, we have the capability to offer a range of standard and customized bioinformatics pipelines to meet the specific needs of customers.
The specific workflow is below.
If you are interested in our services, please feel free to contact us for more details!
References
- Wylezich, C., et al. (2018). "A versatile sample processing workflow for metagenomic pathogen detection." Scientific reports, 8(1), 1-12.
- Plyusnin, I., et al. (2020). "Novel NGS pipeline for virus discovery from a wide spectrum of hosts and sample types." Virus evolution, 6(2), veaa091.
* For research use only.

 Sketch of the basic idea of a "one serves all" analytical framework. (Wylezich, C., et al., 2018).
Sketch of the basic idea of a "one serves all" analytical framework. (Wylezich, C., et al., 2018).