Metabolomics in Influenza Virus Infection
Influenza viruses are common infectious pathogens that pose a threat to avian and mammalian species. The virus continues to evolve, causing seasonal epidemics that lead to significant morbidity and mortality. Despite the annual vaccination program and the use of antiviral drugs, the influenza virus infection and its various complications (e.g. chronic lung disease and asthma) are yet to be adequately addressed. As a result, it is essential to elucidate its pathogenesis and find novel treatment strategies. In addition to proteomics, metabolomics has been applied to the study of influenza virus infection, involving the pathogenesis of the virus, the development of novel therapies, and the diagnosis and prognosis of patients with influenza virus infection.
Metabolic perturbations in influenza infection
Influenza viruses with single-stranded, negative-sense, and segmental RNA genomes are members of the Orthomyxoviridae family. Influenza viruses are characterized by rapid evolution and can cause respiratory infections in mammals (e.g., humans, pigs, horses) and birds (e.g., chickens, waterfowl). To elucidate influenza pathogenesis and find new treatment methods, it is necessary to understand metabolic alterations required for influenza virus infection. According to available metabolomics studies, influenza viruses take advantage of various cellular metabolic pathways to replicate and produce viral particles, involving glucose, lipid, amino acid metabolism, and so on. In turn, the immune response to influenza infection also affects metabolic pathways. Interferon (IFN) is one of these mediators that acts on several metabolites (including nitric oxide, NO, and indoleamine-2,3-dioxygenase, IDO). Following viral infection, IFN production affects cell function by altering membrane composition, amino acid synthesis, and lipid metabolism.
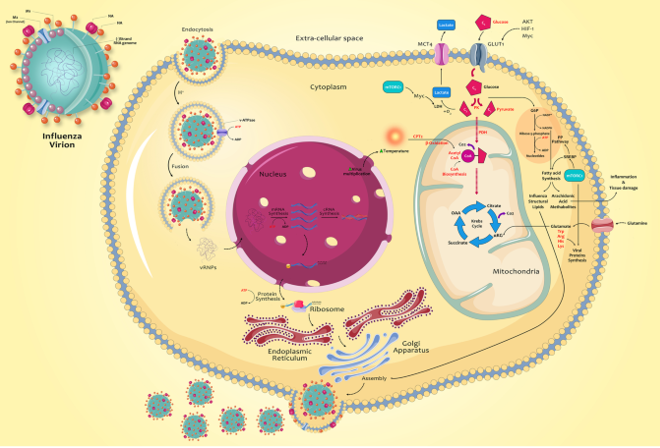 Fig1. Metabolic changes caused by influenza infection and related mechanisms. (Keshavarz, M., et al, 2020)
Fig1. Metabolic changes caused by influenza infection and related mechanisms. (Keshavarz, M., et al, 2020)
Metabolomics and influenza virus
Novel therapeutic approaches by targeting metabolic pathways
Since influenza virus causes seasonal epidemics that lead to significant morbidity and mortality, more attention has been paid to the prevention and treatment of the disease. At present, there are mainly three groups of therapeutic drugs for influenza infection, including M2 inhibitors, polymerase inhibitors, and neuraminidase inhibitors. However, antiviral drug resistance in influenza infection has become a global problem in recent years. There is an urgent need to develop novel antiviral drugs. One of the promising strategies aimed at preventing influenza infection is based on targeted inhibition of these cellular metabolic pathways. Here, we list several key metabolic pathways associated with influenza virus infection, including,
- PI3K/mTOR signaling pathway and PI3K-AKT-mTOR pathway
- The levels of ATP and pyruvate dehydrogenase (PDH)
- The sterol metabolism pathway
- The SREBP-linked pathways
Metabolomics for the diagnosis and prognosis of influenza virus infection
H1N1 influenza infection is a major health burden and can be life-threatening, especially in the elderly and patients with comorbid diseases. In adults, pneumonia induced by influenza has a relatively high mortality. Using nontargeted proton nuclear magnetic resonance (1H-NMR) and gas chromatography-mass spectrometry (GC-MS) approaches, researchers found that plasma-based metabolomics can be used for the early diagnosis and prognosis of H1N1 influenza pneumonia. The metabolic profile of H1N1 infection has a distinct signature compared to bacterial infection and ventilated intensive care unit (ICU) controls, which can be exploited for diagnostic purposes. Moreover, as a highly sensitive and specific tool, metabolomics can also be used for the 90-day prognosis of mortality in H1N1 pneumonia. In addition, a recent study using statistical models and machine learning models demonstrated that an untargeted metabolomics approach from nasopharyngeal samples can be applied for the diagnosis of influenza infection by identifying distinct metabolic signatures.
Metabolomics analysis is a new technology to investigate the metabolites produced by or altered in an organism, which provides a valuable approach for the study of complex diseases. Creative Proteomics has been developing metabolomics detection methods and data analysis methods for many years. Based on professional scientists and advanced platforms, we can accelerate our customers' projects to the next level. For more information on how we can help you, please feel free to contact us.
References
- Banoei, M. M., et al. (2017). "Plasma metabolomics for the diagnosis and prognosis of H1N1 influenza pneumonia." Critical Care, 21(1), 1-15.
- Hogan, C. A., et al. (2021). "Nasopharyngeal metabolomics and machine learning approach for the diagnosis of influenza." EBioMedicine, 71, 103546.
- Keshavarz, M., et al. (2020). "Metabolic host response and therapeutic approaches to influenza infection." Cellular & molecular biology letters, 25(1), 1-19.
Related services
* For research use only.

 Fig1. Metabolic changes caused by influenza infection and related mechanisms. (Keshavarz, M., et al, 2020)
Fig1. Metabolic changes caused by influenza infection and related mechanisms. (Keshavarz, M., et al, 2020)