Multi-Omics in Rabies Virus Infection
Rabies is an invariably fatal neurological disease, which seriously threatens human and animal health. Once clinical symptoms are apparent, the results are nearly always fatal. However, existing treatments are only partially effective. In order to achieve improved treatment options as well as the eventual elimination of the disease, a thorough understanding of rabies virus (RABV) pathogenesis is necessary. Recently, multi-omics technology has emerged as a powerful tool in diverse fields of medical research, including understanding viral pathogenesis. Although multi-omics data on RABV are not yet available, existing omics studies such as genomics, transcriptomics, proteomics and metabolomics could also contribute to the current understanding of the pathogenesis of rabies.
Rabies and rabies virus (RABV)
Rabies are caused by the neurotropic rabies virus (RABV) and a variety of mammalian vectors carry this virus, including humans, dogs, foxes, raccoons, and bats. About 95 percent of human infections are caused by dog bites. There is an incubation phase after exposure and followed by the development of influenza-like symptoms and severe neurotropic symptoms. People with rabies encephalitis often die within a few weeks. RABV, belonging to the Rhabdoviridae family and the Lyssavirus genus, is a bullet-shaped enveloped virus. The virus has a relatively simple RNA genome that encodes five proteins: nucleoprotein (N), glycoprotein (G), phosphoprotein (P), RNA polymerase (L), and matrix protein (M). These proteins play an important role in viral replication and pathogenesis.
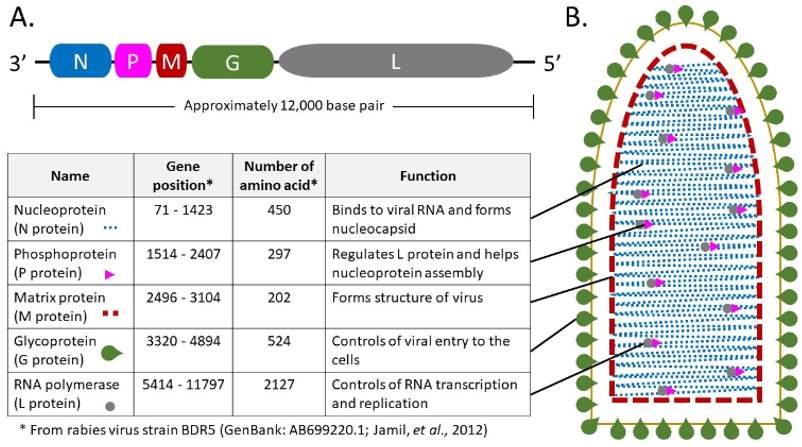 Fig1. RABV genome organization and protein localization in RABV particles. (Chienwichai, P., & Reamtong, O., 2020)
Fig1. RABV genome organization and protein localization in RABV particles. (Chienwichai, P., & Reamtong, O., 2020)
Application of multi-omics to RABV
Multi-omics studies of RABV pathogenesis
Genome sequencing is a useful tool for studying the genetic variation of viruses. The pathogenesis of RABV, such as identifying links between viral genome characteristics and clinical manifestations of disease can be addressed using genomic approaches. So far, there are several genomic studies of RABV, involving viral pathogenesis and phylogenetic analyses. Transcriptomic approaches have been used widely to investigate RABV infection both in vitro and in vivo. Transcriptome studies have found that there are two major mechanisms of RABV pathogenesis, including immune responses and alteration of neuronal homeostasis. Transcriptomic approaches have also been employed to identify potential markers of RABV infection. Proteomic studies and bioinformatic analysis have showed that RABV infection leads to dysregulation of cytoskeletal and structural proteins, thus resulting in impaired integrity and cellular structure. Moreover, RABV-induced autophagy has been suggested as another mechanism of neuronal pathology. Proteomic methods have been used to study RABV pathogenesis and discover novel markers of RABV infection. Post-translational modification (PTM) plays an important role in understanding protein function and cell biology. However, there have been no studies on PTM of the proteome in RABV infection. Therefore, the relationship between PTM of host proteins and the pathogenesis of RABV requires further study. In addition, a few metabolic studies of RABV infection have been performed.
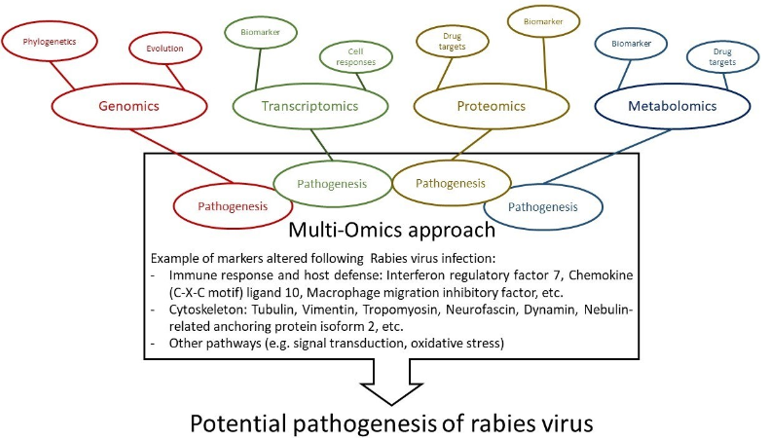 Fig2. Schematic diagram of multi-omics approaches to understand rabies pathogenesis. (Chienwichai, P., & Reamtong, O., 2020)
Fig2. Schematic diagram of multi-omics approaches to understand rabies pathogenesis. (Chienwichai, P., & Reamtong, O., 2020)
Potential pathways involved in rabies pathogenesis identified using multi-omics techniques
Omics research findings come from data-driven processes that can help other researchers target key molecules or pathways. Several potential pathways involved in the pathogenesis of rabies have been identified using multi-omics techniques, including:
- Immune response and host defense
- Alteration of cytoskeletal proteins
- Impairment of signal transduction
- Oxidative stress
- Alteration of cellular metabolism
- Disturbance of transcription and translation factors
- Autophagy and death-mediated factors
- Neuronal dysfunction
- Increased infectivity conferred by viral genome mutations
Creative Proteomics is able to provide transcriptomics, proteomics and metabolomics experiments and multi-omics joint analysis services. Thanks to our long-term experience, high degree of expertise in virus research, and the application of modern technologies, we can provide high-quality and customized service to greatly accelerate our customers’ project progress and success. For more information on how we can help you, please feel free to contact us. We’ll get back to you in time.
Reference
- Chienwichai, P., & Reamtong, O. (2020). “Application of multi-omics technologies to decipher rabies pathogenesis.” The Thai Journal of Veterinary Medicine, 50(2), 129-136.
Related services
* For research use only.

 Fig1. RABV genome organization and protein localization in RABV particles. (Chienwichai, P., & Reamtong, O., 2020)
Fig1. RABV genome organization and protein localization in RABV particles. (Chienwichai, P., & Reamtong, O., 2020) Fig2. Schematic diagram of multi-omics approaches to understand rabies pathogenesis. (Chienwichai, P., & Reamtong, O., 2020)
Fig2. Schematic diagram of multi-omics approaches to understand rabies pathogenesis. (Chienwichai, P., & Reamtong, O., 2020)