Viral Single-Cell Metabolomics Service
Metabolomics has become one of the prime areas in virology research. Although valuable information regarding viral replication and modulation of host metabolic pathways has been extracted, the results are often not representative of the real situations because of the herd effects of infected cells. Creative Proteomics is an international contract research organization (CRO). We continue to expand our services and improve our existing resources to help scientists accelerate their research and support their projects. We have launched a single cell metabolomics (SCM) platform based on precise sampling methods accompanied by robust analytical techniques. On our platform, we can help our clients investigate the cellular heterogeneity as well as differentiation of infected cells from healthy cells, providing deeper insights into the virus-host interactions.
Single-cell metabolomics (SCM) and virus research
Metabolomics analysis contributes to the understanding of host-virus interactions by generating immediate information on the ultimate fates of the analytes and the exact downstream effects. It can provide a shot of dynamic and vulnerable host metabolites in response to specific stages of viral regulation, playing important roles in determining checkpoints and therapeutic targets for effective anti-viral mechanisms. Single cell metabolomics (SCM) is significantly superior to population studies. It can provide specific temporal information in terms of cell differentiation and division, cell communication, stress responses, and interaction with surroundings during viral infection. So far, SCM techniques have yet to make a significant contribution to understanding virus-host interactions. However, with the rapid development in this field, we believe that these techniques will greatly help scientists to explore the altering pattern of viral interactions in cellular heterogeneity under variable host subjects and ecosystems.
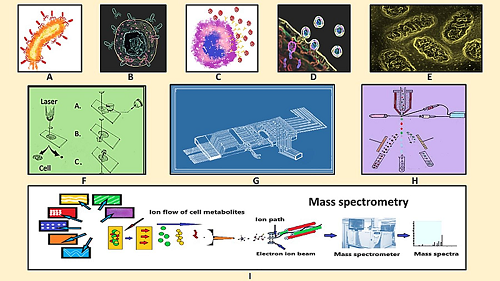 Fig1. Schematic diagram of virus-host interaction, single cell isolation and single cell metabolomics analysis. (A) Virus infecting the host cell, (B) virus-macrophage interaction, (C) virus encounters immune cell, (D) virus-host cell interaction, (E) metabolic changes at sub-cellular level, (F) Single cell isolation by laser microdissection, (G) microfluidic chip for single cell isolation, (H) fluorescence-based cell sorting, and (I) mass spectrometry for single cell metabolomics analysis. (Kumar, R., et al, 2020)
Fig1. Schematic diagram of virus-host interaction, single cell isolation and single cell metabolomics analysis. (A) Virus infecting the host cell, (B) virus-macrophage interaction, (C) virus encounters immune cell, (D) virus-host cell interaction, (E) metabolic changes at sub-cellular level, (F) Single cell isolation by laser microdissection, (G) microfluidic chip for single cell isolation, (H) fluorescence-based cell sorting, and (I) mass spectrometry for single cell metabolomics analysis. (Kumar, R., et al, 2020)
Single-cell metabolomics analytical services
Sample preparation
The isolation of a single target cell is a key step in SCM analysis, which directly reflects the precision of SCM data of the downstream analytical platforms. We employ popular automatic high throughput single-cell isolation techniques employed for SCM analysis, including fluorescence-based cell sorting (FACS) and magnetic-based cell sorting (MACS). In addition, capillary electrophoresis (CE) has the characteristics of fast, high efficiency, high resolution, good reproducibility, and easy automation. This technique can be combined with mass spectrometry (MS) to achieve single-cell metabolic analysis.
Technical platform
- AB SCIEX QTRAP 6500+
Mass range (m/z): 5~2000
- AB SCIEX Triple TOF 6600+
Mass range (m/z): 5-40,000 amu
- AB SCIEX CESI 8000+
Applications of our metabolomics analysis
- The metabolite profiles of virus infection
- Viral disease diagnosis
- New biomarker and drug discovery
- Study of metabolic pathways as well as their perturbations during viral infection
For more information on how we can help you, please feel free to contact us. We look forward to working with you on your next project.
Reference
- Kumar, R., et al. (2020). "Single cell metabolomics: a future tool to unmask cellular heterogeneity and virus-host interaction in context of emerging viral diseases." Frontiers in Microbiology, 11, 1152.
* For research use only.

 Fig1. Schematic diagram of virus-host interaction, single cell isolation and single cell metabolomics analysis. (A) Virus infecting the host cell, (B) virus-macrophage interaction, (C) virus encounters immune cell, (D) virus-host cell interaction, (E) metabolic changes at sub-cellular level, (F) Single cell isolation by laser microdissection, (G) microfluidic chip for single cell isolation, (H) fluorescence-based cell sorting, and (I) mass spectrometry for single cell metabolomics analysis. (Kumar, R., et al, 2020)
Fig1. Schematic diagram of virus-host interaction, single cell isolation and single cell metabolomics analysis. (A) Virus infecting the host cell, (B) virus-macrophage interaction, (C) virus encounters immune cell, (D) virus-host cell interaction, (E) metabolic changes at sub-cellular level, (F) Single cell isolation by laser microdissection, (G) microfluidic chip for single cell isolation, (H) fluorescence-based cell sorting, and (I) mass spectrometry for single cell metabolomics analysis. (Kumar, R., et al, 2020)