Proteomics in HSV-1 Research
Herpes simplex virus (HSV) is ubiquitous in the human population (approximately two-thirds of the world's population are infected) and has the ability to establish lifelong infections. Similar to other herpesviruses, HSV encodes a series of proteins, including proteins associated with viral DNA replication, efficient viral gene expression as well as transcription and translation of cellular genes. These virally expressed proteins not only exhibit multiple different functions but are also associated with a variety of cellular and viral proteins. In order to understand the function of viral proteins and the biology of HSV, proteomics approaches have been applied. We focus on herpes simplex virus 1 (HSV-1), an extensively studied herpesvirus that causes a variety of human diseases, such as encephalitis, mucocutaneous, keratitis, and skin diseases. A recent proteomics study of HSV-1 will be reviewed here.
Herpes simplex virus 1 (HSV-1)
HSV-1 is a widespread double-stranded DNA (dsDNA) virus that can lead to a contagious and persistent infection in most human populations. HSV-1 is able to establish lifelong infections with reactivation triggered by stimuli, such as tissue damage and compromised immunity. These infections generally range from asymptomatic, mild symptoms, such as oral sores and watery blisters on the skin or mucous membranes, to severe diseases, such as herpes keratitis and herpes encephalitis. In general, HSV-1 initially infects epithelial cells in the form of a lytic infection and then enters peripheral neurons to establish latency. HSV-1 exploits various host cell systems to promote their own replication. So far, there are at least 84 different viral proteins encoded by HSV-1.
Proteomic study of HSV-1
Time-resolved global and chromatin proteomics analysis of HSV-1 infection
A early as 2017, a study conducted a large-scale proteomic analysis of HSV-1 infection. The study was performed in human foreskin fibroblast (HFF) cells and presented a system-level characterization of proteome dynamics during infection through a multi-dimensional analysis, including viral proteome identification and quantification, host proteome identification and quantification, chromatin bound proteomes, phosphoproteomes, and post-translational modifications (PTMs) on cellular histones. In this work, researchers characterized changes in protein post-translational modification following infection and quantified the abundance of approximately 4,000 host proteins, indicating that the proteome composition of the chromatin of HFF cells is highly affected during HSV-1 infection and there are numerous phosphorylation events on viral proteins.
A subcellular quantitative proteomic analysis of HSV-1 infection
The mechanisms of HSV-1 infection as well as HSV-1's interactions with various host cells have been studied extensively. A stable isotope-labeled amino acid culture (SILAC)-based quantitative proteomics method was applied to explore the proteomic effects of HSV-1 infection on HEK 293T cells. In the study, a total of 6607 host proteins and 498 differential proteins were identified using a subcellular fractionation strategy and high-performance mass spectrometry.
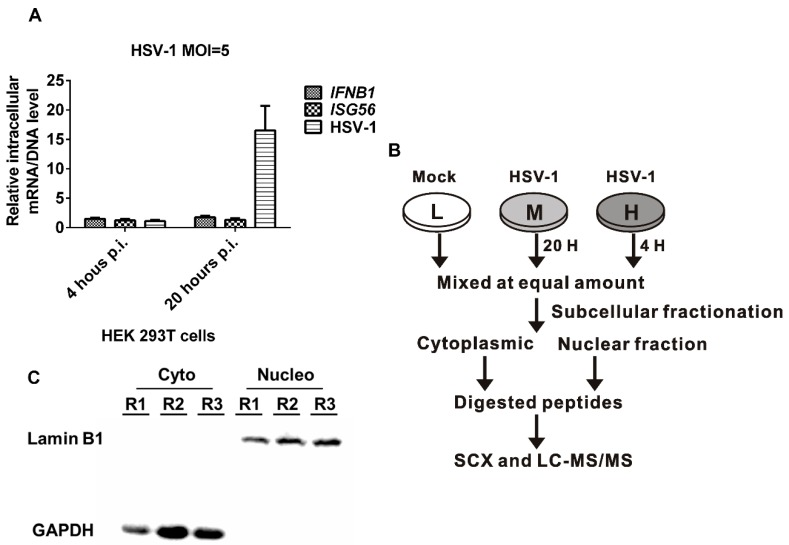 Fig1. Subcellular quantitative proteomic analysis of herpes simplex virus type 1 (HSV-1)-infected HEK 293T cells.(Wan, W., et al, 2019)
Fig1. Subcellular quantitative proteomic analysis of herpes simplex virus type 1 (HSV-1)-infected HEK 293T cells.(Wan, W., et al, 2019)
Temporal proteomics analysis of HSV-1 infection
To further characterize the molecular mechanisms of host and viral proteome changes during HSV-1 infection, researchers employed quantitative temporal viromics (QTV) to investigate a single replication cycle of HSV-1 in human keratinocytes. QTV is a highly multiplexed method that allows the analysis of temporal changes in host and viral proteins during productive infections. The method is based on tandem mass spectrometry (TMTs) and tertiary mass spectrometry (MS3), which can accurately quantify each protein. In the study, researchers had identified several host-cell proteins that can be rapidly degraded by HSV-1, as well as a specific mediated pathway. The research highlights an efficient mechanism to manipulate the abundance of host proteins on the surface of infected cells, namely, HSV-1 specifically targets a cellular trafficking factor.
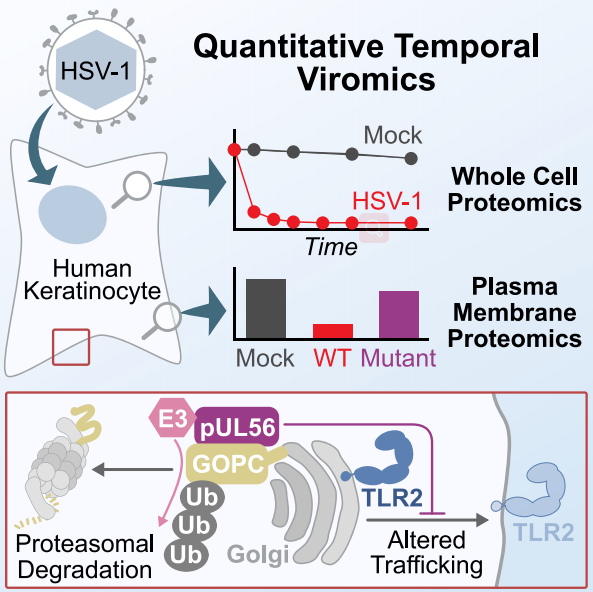 Fig2. Temporal proteomic analysis of herpes simplex virus 1 infection reveals cell-surface remodeling via pUL56-mediated GOPC degradation. (Soh, T. K.,et al, 2020)
Fig2. Temporal proteomic analysis of herpes simplex virus 1 infection reveals cell-surface remodeling via pUL56-mediated GOPC degradation. (Soh, T. K.,et al, 2020)
Creative Proteomics provides a series of proteomic techniques to help customers quickly and comprehensively investigate viral proteomics. For more information on how we can help you, please feel free to contact us or directly send us an inquiry.
References
- Kulej, K., et al. (2017). "Time-resolved global and chromatin proteomics during herpes simplex virus type 1 (HSV-1) infection." Molecular & Cellular Proteomics, 16(4), S92-S107.
- Wan, W., et al. (2019). "A subcellular quantitative proteomic analysis of herpes simplex virus type 1-infected HEK 293T cells." Molecules, 24(23), 4215.
- Soh, T. K.,et al. (2020). "Temporal proteomic analysis of herpes simplex virus 1 infection reveals cell-surface remodeling via pUL56-mediated GOPC degradation." Cell reports, 33(1), 108235.
Related services
* For research use only.

 Fig1. Subcellular quantitative proteomic analysis of herpes simplex virus type 1 (HSV-1)-infected HEK 293T cells.(Wan, W., et al, 2019)
Fig1. Subcellular quantitative proteomic analysis of herpes simplex virus type 1 (HSV-1)-infected HEK 293T cells.(Wan, W., et al, 2019)  Fig2. Temporal proteomic analysis of herpes simplex virus 1 infection reveals cell-surface remodeling via pUL56-mediated GOPC degradation. (Soh, T. K.,et al, 2020)
Fig2. Temporal proteomic analysis of herpes simplex virus 1 infection reveals cell-surface remodeling via pUL56-mediated GOPC degradation. (Soh, T. K.,et al, 2020)