Viral HDX-MS Analysis Service
The construction of capsid surrounding the viral genetic material is an important part of virology research. There are three main functions of viral protein capsid, including protecting the genetic material from digestive enzymes, assisting the virus to attach to the host cell membrane, and transmitting the genetic information into the host cells. Creative Proteomics is a leading provider in the field of viral proteomics services. In order to investigate the structure and conformational dynamics of capsids, we are proud to offer hydrogen-deuterium exchange mass spectrometry (HDX-MS) experiment. We are dedicated to helping our customers understand the specific function of these structures in the virus lifecycle and further fight against viral infection.
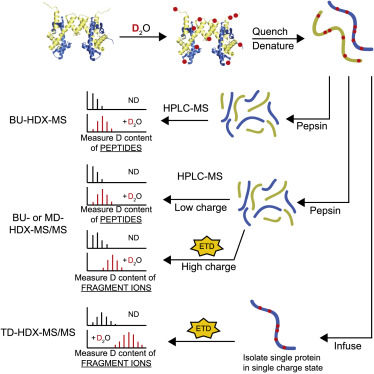 Schematic of HDX-MS Experiments. BU-HDX-MS: bottom-up HDX-MS; MD-HDX-MS/MS: middle-down HDX-MS/MS; TD-HDX- MS/MS:top-down HDX-MS/MS (Karch, K. R., et al, 2018)
Schematic of HDX-MS Experiments. BU-HDX-MS: bottom-up HDX-MS; MD-HDX-MS/MS: middle-down HDX-MS/MS; TD-HDX- MS/MS:top-down HDX-MS/MS (Karch, K. R., et al, 2018)
Elucidating protein conformation, structure, and dynamics with HDX-MS
As an isotope labeling and integrated structural biology technique, HDX-MS has been used in many protein-based systems. The principle of HDX-MS is measuring the rate of hydrogen-deuterium exchange at specific sites of interest, thereby the approach can be applied to infer information on protein structure, such as protein-protein interaction (PPI) sites, allosteric effects, as well as conformational changes caused by post-translational modifications (PTMs).
The main types of information obtained using HDX-MS
- Conformations of proteins and protein complexes.
- Protein dynamics
- Biomolecule binding
- Allosteric effects
HDX-MS analysis in virology research
In the past years, HDX-MS has also been applied to investigate the structural dynamics of viruses throughout their replication cycle, ranging from host–cell binding, membrane fusion, to genome replication and transcription, nucleocapsid assembly, and budding of new virions. In HDX-MS experiments, the targeted viral proteins are firstly transferred into deuterated "heavy" water. Certain protons derived from the protein freely exchange with deuterons in a deuterated buffer. Then, the kinetics and extent of deuterons exchanging with hydrogen on different functional groups of the protein are monitored by MS. Importantly, HDX-MS is not limited by the size of proteins or protein complexes, which is a key advantage of HDX-MS over other integrated structural biology methods. Thus, the method is widely used for detecting virus structure. So far, HDX-MS has been successfully applied to investigate capsid assembly and maturation of the HIV-1 virus, the capsid protein dynamics of the hepatitis B virus, and so on.
Service offering in Creative Proteomics
The accumulated improvements in sample preparation, LC-MS, automation, and informatics technologies have made HDX-MS methods much more widely accessible throughout academia and the biopharmaceutical industry. Based on advanced instruments, professional scientists, and robust workflow, we can conduct HDX-MS analysis to determine local conformational dynamics as well as gain insight into virus assembly mechanisms and capsid maturity. Our customized services can accelerate the progress of our client's studies, including,
- Virial biophysical characterization
HDX-MS can be applied for biophysical characterization of virus particles, such as the response of the virus particle to chemical denaturation, pH, and changes in temperature.
- Studying the viral replication cycle, such as virus-host recognition and cell entry
The ability of HDX-MS to detect multiple co-occurring conformational states and to probe structural dynamics makes it suitable for studying the typical pre-fusion and post-fusion conformational changes of viral envelope glycoproteins.
- The development of vaccines and antiviral drugs
The epitope mapping experiments with HDX-MS can be used to understand the mechanisms of neutralizing immunity when the host response to viral infections as well as screen and optimize these monoclonal antibodies for therapeutic applications.
If you are interested in our services, please feel free to contact us. We look forward to providing services for your next project.
Reference
- Karch, K. R., et al. (2018). "Hydrogen-deuterium exchange coupled to top-and middle-down mass spectrometry reveals histone tail dynamics before and after nucleosome assembly." Structure, 26(12), 1651-1663.
* For research use only.

 Schematic of HDX-MS Experiments. BU-HDX-MS: bottom-up HDX-MS; MD-HDX-MS/MS: middle-down HDX-MS/MS; TD-HDX- MS/MS:top-down HDX-MS/MS (Karch, K. R., et al, 2018)
Schematic of HDX-MS Experiments. BU-HDX-MS: bottom-up HDX-MS; MD-HDX-MS/MS: middle-down HDX-MS/MS; TD-HDX- MS/MS:top-down HDX-MS/MS (Karch, K. R., et al, 2018)