Mass Spectrometry-Based Viral Proteomics
 Viruses are parasites of living organisms. They replicate and proliferate in host cells and continuously evolve to modulate and adjust to the harsh host environment. As a consequence of viral infection, a series of proteome changes are triggered during the progression of the infection, including alterations in protein abundance and interactions, protein post-translational modifications (PTMs), subcellular localizations, and so on. MS-based proteomics is capable of precisely identifying viral and cellular proteins, enabling us to detect important pathway proteins, enlarging protein-protein interaction maps. Based on the popular and improved MS-based approaches, Creative Proteomics provides custom experimental design and detailed research reports according to customers' specific project needs, promoting the investigations of viral proteomes and host responses associated with viral infections.
Viruses are parasites of living organisms. They replicate and proliferate in host cells and continuously evolve to modulate and adjust to the harsh host environment. As a consequence of viral infection, a series of proteome changes are triggered during the progression of the infection, including alterations in protein abundance and interactions, protein post-translational modifications (PTMs), subcellular localizations, and so on. MS-based proteomics is capable of precisely identifying viral and cellular proteins, enabling us to detect important pathway proteins, enlarging protein-protein interaction maps. Based on the popular and improved MS-based approaches, Creative Proteomics provides custom experimental design and detailed research reports according to customers' specific project needs, promoting the investigations of viral proteomes and host responses associated with viral infections.
Applications of proteomics platforms on virus research
Mass spectrometry (MS)-based proteomics is one of the cutting-edge molecular technologies and has emerged as a powerful biotechnological tool in the post-genomic era. The main benefit of MS-based technology in proteomics is capable of providing a wide range of proteomic information in a dynamic and quantitative manner, with multiplexity, high-throughput capability, and high reproducibility. MS-based proteomic platforms have been successfully used to better understand the mechanisms of viral infection. The main applications of proteomics platforms in virus research include,
- Offer the global expression pattern of thousands of proteins and the potential pathways regulated in the cellular system during the progression of the infection.
- Identify the virus-host protein interactions.
- Identify post-translational modifications of viral protein and host cell proteome.
- Provide a highly repeatable multiplexed diagnostic tool for viruses.
Service offering
An accurate understanding of viral structures and functions demands reliable techniques for protein characterization. Creative Proteomics is a forward-looking research institute as well as a leading custom service provider in the field of viral proteomics. Based on our experienced scientists and advanced instruments, we are able to provide global customers with several popular MS-based methods for studying viruses and their hosts. Importantly, each method has its own uniqueness and advantages. In addition to offering the most effective MS strategies, we can also provide complementary characterization strategies according to customers' specific project needs.
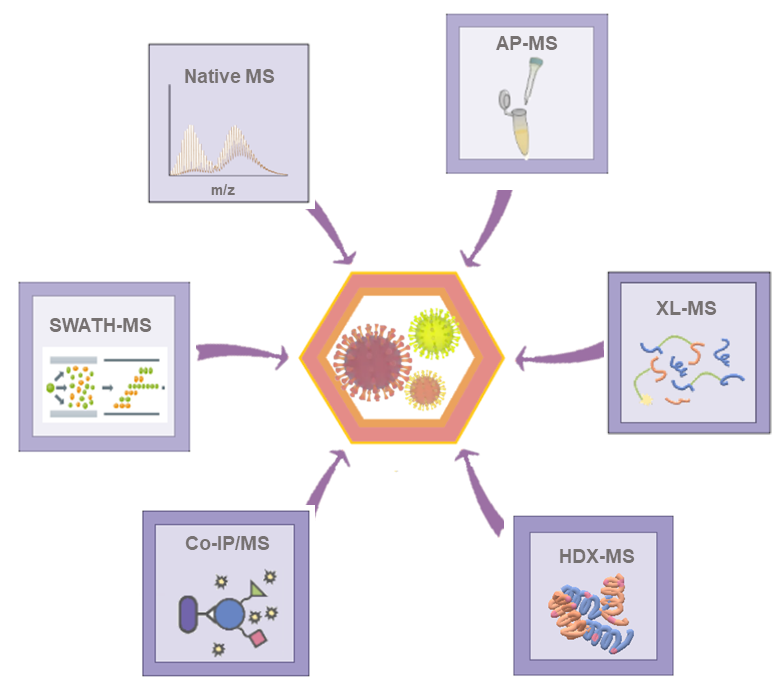
Benefits of our services
- An optimization & scale-up platform
- Professional technical support
- A rapid and tailored solution for specific research applications
- Complete analytical study reports
In order to rapidly respond to harmful new viruses and to develop new antiviral drugs, a detailed and reliable understanding of the structure and function of viruses is essential. MS has emerged as a powerful tool for elucidating virus structure and function. We offer a series of MS techniques together with ongoing improvements, helping our customers obtain more useful information more quickly, and ultimately accelerating the development of vaccines and therapeutics.
If you are unable to find the specific service you are looking for, please feel free to contact us.
Reference
- Ahsan, N., et al. (2021). "Mass spectrometry‐based proteomic platforms for better understanding of SARS-CoV-2 induced pathogenesis and potential diagnostic approaches." Proteomics, 21(10), 2000279.
* For research use only.

 Viruses are parasites of living organisms. They replicate and proliferate in host cells and continuously evolve to modulate and adjust to the harsh host environment. As a consequence of viral infection, a series of proteome changes are triggered during the progression of the infection, including alterations in protein abundance and interactions, protein post-translational modifications (PTMs), subcellular localizations, and so on. MS-based proteomics is capable of precisely identifying viral and cellular proteins, enabling us to detect important pathway proteins, enlarging protein-protein interaction maps. Based on the popular and improved MS-based approaches, Creative Proteomics provides custom experimental design and detailed research reports according to customers' specific project needs, promoting the investigations of viral proteomes and host responses associated with viral infections.
Viruses are parasites of living organisms. They replicate and proliferate in host cells and continuously evolve to modulate and adjust to the harsh host environment. As a consequence of viral infection, a series of proteome changes are triggered during the progression of the infection, including alterations in protein abundance and interactions, protein post-translational modifications (PTMs), subcellular localizations, and so on. MS-based proteomics is capable of precisely identifying viral and cellular proteins, enabling us to detect important pathway proteins, enlarging protein-protein interaction maps. Based on the popular and improved MS-based approaches, Creative Proteomics provides custom experimental design and detailed research reports according to customers' specific project needs, promoting the investigations of viral proteomes and host responses associated with viral infections. 