Multi-Omics in Pathogenic Mosquito-Borne Virus Infection
Pathogenic mosquito-borne viruses, such as Zika virus (ZIKV), Chikungunya virus (CHIV), dengue virus (DENV), West Nile virus (WNV), and Japanese encephalitis virus (JEV), are increasingly becoming a serious public health issue. To better understand mosquito-borne infections and develop new treatments to combat outbreaks of these pathogens, multiple-omics analyses (including transcriptomics, proteomics, and interactomics) and network projection analyses are applied. Creative Proteomics is able to provide transcriptomics, proteomics, and metabolomics experiments and multi-omics joint analysis services. Based on well-established longitudinal cohorts and multi-omics technologies, we can greatly promote the study of many viruses.
Mosquito-borne virus infection
Mosquito-borne infections are widespread, and combinations of these pathogens can lead to a lot of human suffering. Although mosquito-borne diseases have a low mortality rate compared to infectious diseases like Ebola, Congo's Crimea, and Marburg, over 50 billion people have died from mosquito-borne diseases and more than 390 million people are still infected every year. These pathogenic mosquito-borne viruses, including ZIKV, CHIV, DENV, WNV, and JEV, can cause a range of symptoms, such as hemorrhagic fever and encephalitis. Current trends suggest that mosquito-borne diseases have become a serious problem, with greater numbers of infected people and more severe symptoms. Drug repositioning/repurposing is cost-effective and has limited side effects, making it particularly suitable for treating these neglected tropical diseases.
Application of multi-omics in mosquito-borne virus infection
By analyzing GEO datasets (transcriptome data), reviewing the proteome literature (proteome data), and reviewing human-viral PPI literature (protein-protein interaction data), researchers identified signature genes, target proteins, and target pathways associated with DENV, ZIKV, WNV, CHIV, and JEV. According to the results generated from multi-omics analyses, researchers used a network analysis approach to identify potential drug repositioning candidates and classify these drugs. Based on GEO datasets, a connectivity map (CMap) for signature genes was generated. STITCH was used for selecting chemicals that interacted with the identified signature proteins from the proteome and human-viral protein-protein interactions (PPIs). The drug candidates for the five viral infections were identified using the filtering method. Finally, the proteins related to the onset and progression of viral infectious diseases were projected onto the PPI network of the Human Protein Reference Database (HPRD), and drugs for five diseases were classified.
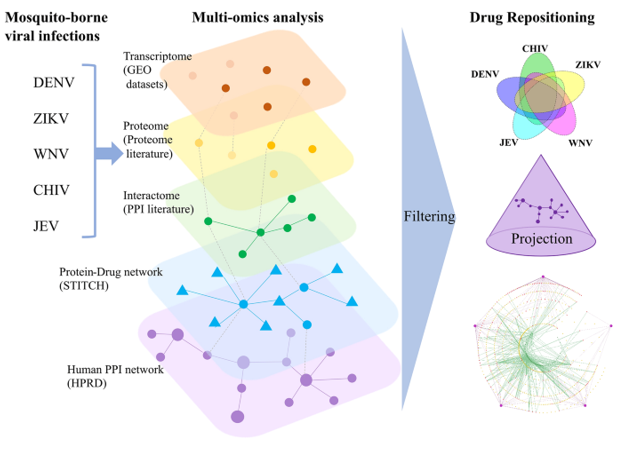 Fig1. Schematic showing the process of drug repositioning against the five viruses (DENV, ZIKV, WNV, CHIV, and JEV). (Amemiya, T., et al, 2021)
Fig1. Schematic showing the process of drug repositioning against the five viruses (DENV, ZIKV, WNV, CHIV, and JEV). (Amemiya, T., et al, 2021)
Based on multi-omics analyses, 77 drug candidates and 146 proteins were identified for the five viral infections. Of these identified candidates, such as valproic acid, resveratrol, paracetamol, and trichostatin A have been reported as effective agents for the treatment of flavivirus-induced diseases.
The combination of multi-omics analyses and network analysis is an effective approach to comprehensively understand the biological process of virus infection and identify novel drug candidates for mosquito-borne virus infection. If you have any needs in this area, please feel free to contact us. We will serve you wholeheartedly.
Reference
- Amemiya, T., et al. (2021). "Application of multiple omics and network projection analyses to drug repositioning for pathogenic mosquito-borne viruses." Scientific reports, 11(1), 1-13.
Related services
* For research use only.

 Fig1. Schematic showing the process of drug repositioning against the five viruses (DENV, ZIKV, WNV, CHIV, and JEV). (Amemiya, T., et al, 2021)
Fig1. Schematic showing the process of drug repositioning against the five viruses (DENV, ZIKV, WNV, CHIV, and JEV). (Amemiya, T., et al, 2021)