Multi-Omics in HCV Infection
Hepatitis C virus (HCV) currently infects over 71 million humans worldwide and is the most common risk factor for hepatocellular carcinoma (HCC) in Western countries. As a major public health threat, the mechanisms of HCV infection, liver disease progression, and hepatocarcinogenesis need to be further studied. Based on multi-omics approaches and data analysis (including genomic, proteomic, and metabolomic analyses) from in vivo and in vitro, the complex interactions of HCV-host, as well as the information of HCV-associated liver disease, have been identified.
Hepatitis C virus (HCV)
Hepatitis C virus (HCV) is a small-enveloped virus belonging to the family of Flaviviridae. Despite constant progress in HCV diagnosis and treatment, HCV infection remains a serious health problem and a major causative agent of chronic liver diseases. HCV can lead to acute and chronic liver infections, which is the main reason liver disease progresses from chronic inflammatory and metabolic disease to fibrosis, cirrhosis, and ultimately HCC. The virus accounts for up to 25% of HCC cases worldwide. In addition, an estimated 1% of the world's population is infected with HCV, most of whom do not realize they have the virus. HCV infection is a suitable model for studying the general mechanisms of immune evasion and disease pathogenesis due to the well-developed strategies for evading antiviral responses and regulating changes in cellular circuitry and metabolic pathways. HCV can be used to study viral and non-viral liver disease.
Multi-omics studies of HCV pathogenesis
In order to investigate virus-host interaction and the impact of HCV on liver disease biology at the systems level, multi-omics approaches are applied, including proteomics, transcriptomics, and metabolomics. Based on the cutting-edge screening technologies combined with state-of-the-art HCV infection model (HCV-infected cells and chimeric mice), the study has revealed virus-induced perturbations of host cell circuits and the temporal proteogenomic atlas of persistent HCV infection.
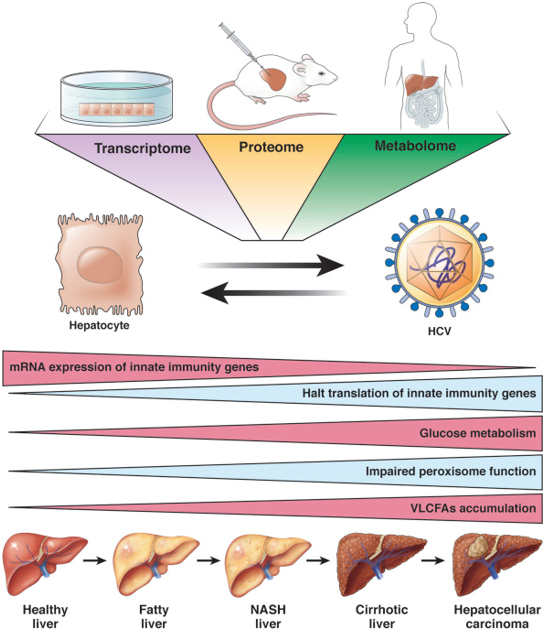 Fig1. Multiomics approach identifies molecular mechanisms leading to liver diseases and HCC. (Gal-Tanamy, M., 2019)
Fig1. Multiomics approach identifies molecular mechanisms leading to liver diseases and HCC. (Gal-Tanamy, M., 2019)
An integrated proteogenomic approach shows the spatiotemporal map of HCV–hepatocyte interactions during infection
There is a multidimensional atlas of persistent HCV infection that reflects a time-resolved proteogenomic state of HCV-infected cells, involving 21,950 mRNAs and 8297 proteins. Through using integrative functional genomic analyses, 90.8% of proteins (7,416 proteins) were uniquely mapped to overlapping mRNAs.
Disparate dynamics between host cell mRNA and proteins upon HCV infection
Differentially expressed genes or proteins were classified according to their representation within a predefined gene set associated with a phenotype using GSEA. The results showed that HCV infection shuts down most non-essential processes at the transcriptional and/or post-transcriptional level, thereby diverting resources to viral replication and persistence. There were several up-regulated pathways, including inflammation, the antiviral response, and proviral signaling pathways.
Pathways identified in response to HCV infection
Based on differential expression, gene set enrichment analyses as well as protein interaction mapping, there are several pathways identified in response to HCV infection. HCV infection perturbs peroxisomal function by increasing glucose metabolism and the STAT3 signaling pathway, which is associated with steatohepatitis and poor prognosis in patients.
Creative Proteomics is able to provide transcriptomics, proteomics, and metabolomics experiments and multi-omics joint analysis services. Thanks to our long-term experience, high degree of expertise in virus research, and the application of modern technologies, we can provide high-quality and customized service to greatly accelerate our customers' project progress and success. For more information on how we can help you, please feel free to contact us. We'll get back to you in time.
References
- Lupberger, J., et al. (2019). "Combined analysis of metabolomes, proteomes, and transcriptomes of hepatitis C virus–infected cells and liver to identify pathways associated with disease development." Gastroenterology, 157(2), 537-551.
- Gal-Tanamy, M. (2019). "Multi-omic Analyses Reveal Complex Interactions Between HCV and Hepatocytes Demonstrating That the Red Queen Is Up and Running." Gastroenterology, 157(2), 300-302.
Related services
* For research use only.

 Fig1. Multiomics approach identifies molecular mechanisms leading to liver diseases and HCC. (Gal-Tanamy, M., 2019)
Fig1. Multiomics approach identifies molecular mechanisms leading to liver diseases and HCC. (Gal-Tanamy, M., 2019)