
- Home
- DNA/RNA Modification LC-MS Analysis Service
LC-MS/MS detection of nucleic acid modifications DNA/RNA modifications are an important aspect of epigenetic modifications, referring to various modifications on DNA, RNA bases A\T\G\C\U, and ribose. Currently, over a dozen DNA modifications and over 100 RNA modifications have been discovered, with methylation modification being the majority. Research on methylation modification is currently widespread and may have significant biological regulatory functions.
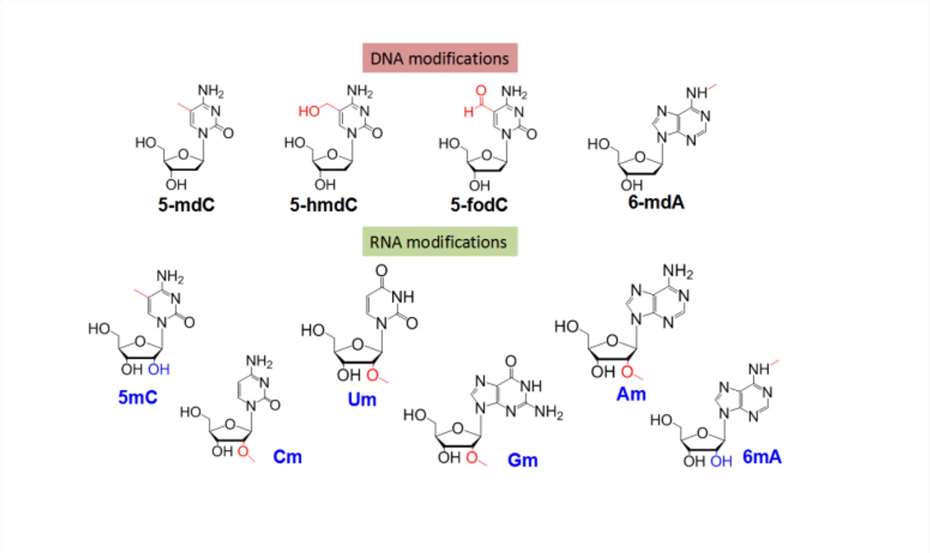 DNA/RNA Modification
DNA/RNA Modification
Creative Proteomics has established a nucleic acid modification analysis method based on the LC-MS/MS platform, enabling the quantitative analysis of four types of DNA modifications (5-mdC, 5-hmdC, 5-fodC, 6-mdA) and nine types of RNA modifications (m5C, m6A, Am, Gm, Cm, Um, m1A, m7G, ac4C). This method provides an ideal technical platform for epigenetic research.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) is capable of detecting highly polar and less stable compounds and enables precise quantification of substances. LC-MS/MS is currently the method that allows for accurate quantification of low-abundance modified bases among all the existing methods for analyzing overall methylation levels. By combining LC-MS/MS with certain sample pre-processing methods, the detection of DNA/RNA modifications in various biological samples can be improved. The workflow for nucleic acid modification analysis is shown in the diagram below:
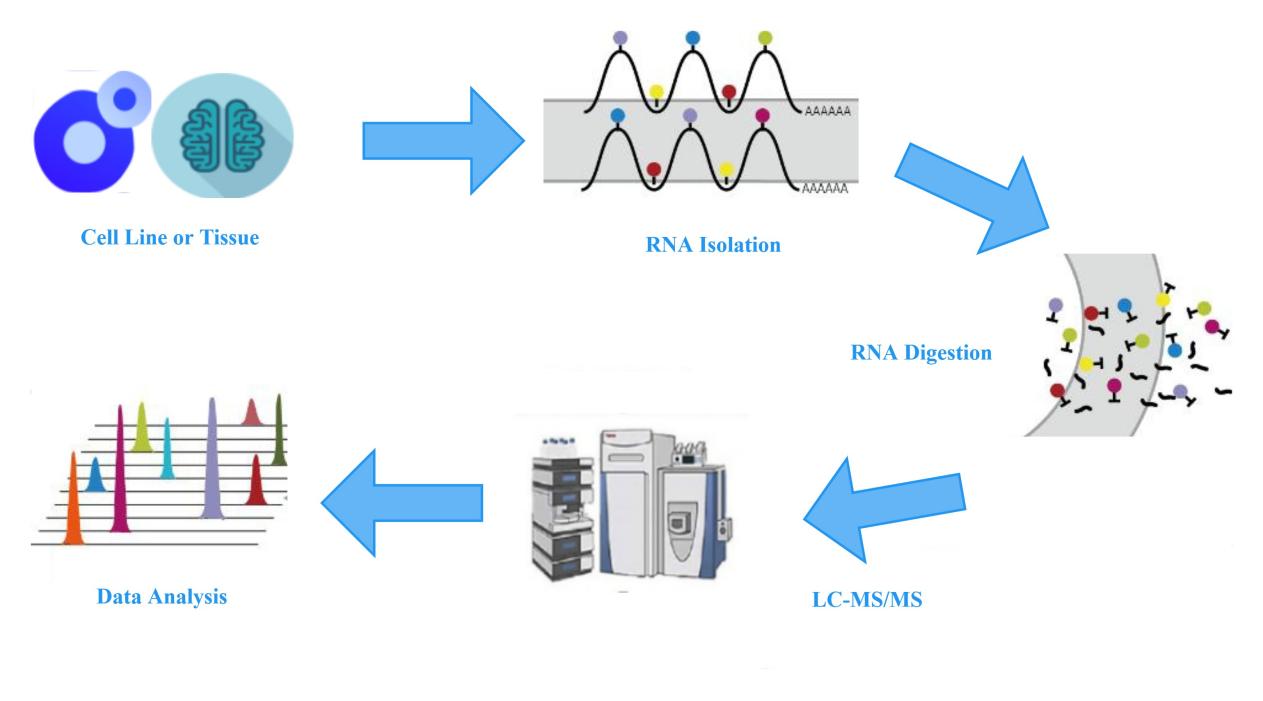
mRNA Modification LC-MS Analysis Service
tRNA Modification LC-MS Analysis Service
m6A Modification LC-MS Analysis Service
The DNA/RNA Modification LC-MS Analysis Service from Creative Proteomics evaluates dozens ofnucleoside alterations and characterizes the global modification profile of every nucleic acid class you select or isolate.
DNA Sample Requirements
| Detection Type | Detection Method | Quality Requirements | Notes |
|---|---|---|---|
| 5-mdC, 5-hmdC, 6-mdA | Nanodrop | Total ≥ 4μg/sample, concentration ≥ 50ng/μL; OD 260/280: 1.8~2.0, OD 260/230 ≥ 1.8 |
1) Dissolve in ultrapure water; 2) If OD 260/280, 260/230 does not meet the standard but the difference is small, and other conditions meet the requirements, and the sample is not in a viscous state, it can be used for nucleic acid detection; 3) To ensure the smooth progress of the project, it is recommended to provide additional sample quantity for library construction or multiple samples. |
| Gel Electrophoresis | No viscosity, no abnormal color, no impurities/RNA contamination, no smearing bands | ||
| 5-fdC | Same as above | Total ≥ 10-20μg/sample, concentration ≥ 100mg/μL; other requirements same as above | |
| 5-mdC, 5-hmdC, 5-fdC, 6-mdA | Same as above | Total ≥ 10-20μg/sample, concentration ≥ 100ng/μL; other requirements same as above |
RNA Sample Requirements
| Detection Type | Detection Method | Quality Requirements | Notes |
|---|---|---|---|
| m5C, m6A, Am, Um, Gm, Cm, m1A, m7G, ac4C | Nanodrop | Total ≥ 2μg/sample, concentration ≥ 50ng/μL; OD 260/280: 1.8~2.0, OD 260/230 ≥ 1.8 | 1) Dissolve in ultrapure water; 2) If OD 260/280, 260/230 does not meet the standard but the difference is small, and other conditions meet the requirements, and the sample is not in a viscous state, it can be used for nucleic acid detection; 3) To ensure the smooth progress of the project, it is recommended to provide additional sample quantity for library construction or multiple samples. |
| Gel Electrophoresis | No genomic contamination, no protein and impurity contamination, no abnormal coloration |
Our products and services are for research use only.