
- Home
- PTMs Proteomics
- PTM Quantitative Analysis
- Label-Free PTM Quantitative Analysis
Although the use of labeling strategies in proteome quantification is effective, it is time-consuming, expensive, and complex in terms of the sample preparation process as well as data analysis. Creative Proteomics is also dedicated to providing novel and accurate label-free quantification approaches for post-translational modification (PTM) analysis. Based on our years of experience in MS and a team of professional MS experts, we have developed a robust workflow combining affinity purification and liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis for quantifying changes of various PTMs.
Label-free quantification is a method for determining the relative amount of protein in two or more biological samples without using stable isotopes to chemically bind and label the protein. These methods are divided into two main strategies. The first method is based on cross-checking the number of MS/MS spectra of protein peptides acquired between different standard samples (spectral counting), and the second method entails measuring and comparing the areas of peptide precursor ion chromatographic peaks between different samples (ion intensities). There are two main advantages of label-free methods, including simpler sample preparation and direct comparison of multiple samples, making them particularly suitable for studies requiring statistical analysis of technical and biological replicates.
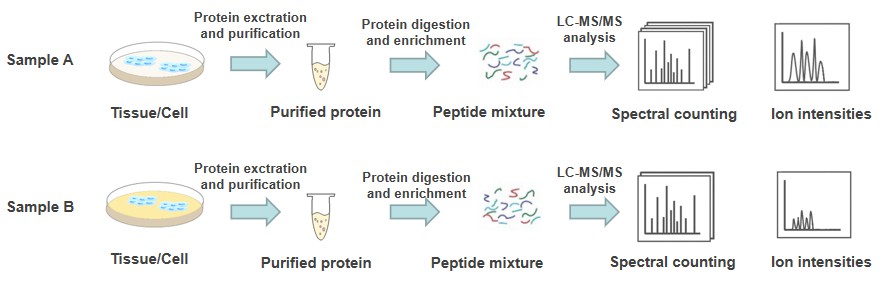
Label-free quantification is a fast, cost-efficient, and easy-handling relative method that has been used to measure differential proteomics expression on a large scale. Combining affinity purification and LC-MS/MS analysis, we developed a robust workflow that is compatible with label-free strategies. During label-free relative quantification, we consider the effects of sample heterogeneity and contaminants in the sample, and we focus on reducing technical variation in the various steps of sample preparation and during LC-MS measurements. A key element of successful label-free quantification is consistent instrument performance throughout the data collection period. Our label-free quantification has been proven the high reproducibility and our workflow can be broadly adopted for quantifying changes of various PTMs. In addition, we have professional PTM bioinformatics experts who provide label-free quantitative data processing services using multiple sources as well as commercial software packages. Our specific services are as follows:

For clients wishing to quantify protein PTMs by MS analysis, Creative Proteomics now offers a range of labeled and unlabeled proteomic-based methods and services. We are committed to employing an application-oriented development strategy, with a particular focus on experimental design and high-quality services. Please feel free to contact us for more details of our services.
References
Our products and services are for research use only.