
- Home
- PTMs Proteomics
- PTM Quantitative Analysis
- iTRAQ/TMT-Based PTM Quantitative Analysis
With advances in mass spectrometry (MS) analysis and post-translational modification (PTM) enrichment techniques, the identification and quantification of hundreds and thousands of PTMs are possible. To help our global customers achieve relative quantification of PTMs across multiple biological samples, Creative Proteomics also provides chemical labeling methods for MS-based quantitative proteomics, including isobaric tag for relative and absolute quantification (iTRAQ) and tandem mass tag (TMT). These chemical labeling techniques allow the comparison and multiplexing of multiple samples in a single MS run.
Recently, MS-based labeling strategies such as SILAC, TMT, and isobaric tag for relative and absolute quantification (iTRAQ), have gained popularity in quantitative proteomics and have also been applied successfully to identify and quantify PTMs. Chemical labeling technologies involve the introduction of stable isotopes on target proteins/peptides for quantitative analyses. Depending on the label used, up to 8 (iTRAQ) or 16 samples (TMT) can now be multiplexed, greatly facilitating the use of a wide range of fractionation and enrichment protocols prior to LC-MS analysis. In the case of reporter ion-based iTRAQ and TMT, the same peptides from different samples are always isobaric after labeling, so even multiplexing 10 samples does not significantly increase sample complexity. Due to the multiplexing capability combined with the isobaric nature of the labels, iTRAQ and TMT are well-suited for studies with limited amounts of available protein per sample/condition.
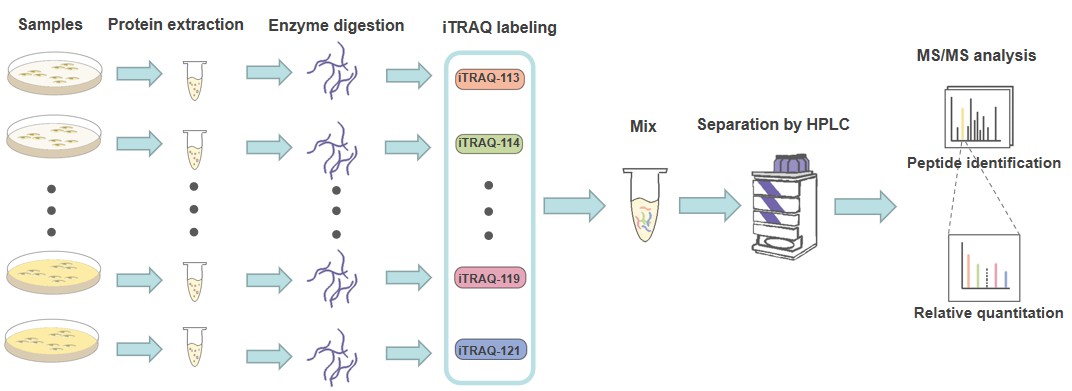
We are dedicated to using a high-sensitivity MS-based proteomics platform based on isobaric labels iTRAQ/TMT to provide a comprehensive and accurate analysis of low-abundance proteins and their PTMs. Overall, we employ quantitative iTRAQ/TMT tag-based proteomics analysis combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) to capture differential protein expression profiles between controls and treatments. Since the labels can alter the physicochemical properties of peptides and directly affect the efficiency of PTM enrichment. Workflow of our service:

Creative Proteomics has an advanced MS platform, years of experience in MS, and a team of professional MS experts. We specialize in a wide range of PTM analyses, including phosphorylation, acetylation, glycosylation, and ubiquitination, among others. For more information about our PTM analysis services and sample requirements, please feel free to contact us.
References
Our products and services are for research use only.