- Home
- Applications
- Protein Post-Translational Modifications in Bacteria
In addition to eukaryotes, protein post-translational modifications (PTMs) are widely found in microbiota, including archaea and bacteria. As one of the important strategies of microbiota for environmental adaptation, protein PTMs control a number of fundamental features of microbial biology by regulating protein complexity. PTMs generally affect enzymatic activity, interaction with partners, protein stability and DNA binding and play a crucial role in microbial cell signaling, gene expression, metabolism, and host-microbe interactions. The study of the role of PTMs in microbial ecology and physiology provides new insights into biological studies from another perspective.
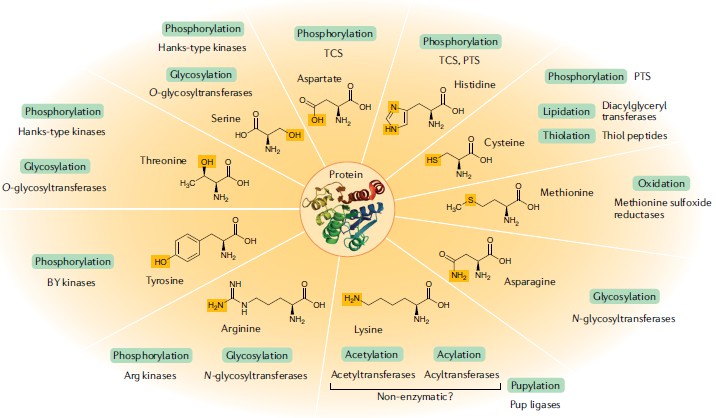 Fig. 1 Protein modifications in bacteria. (Macek, Boris, et al, 2019)
Fig. 1 Protein modifications in bacteria. (Macek, Boris, et al, 2019)
In the last decade, with the advent of advanced protein PTMs detection methods such as modified peptide enrichment combined with high-accuracy mass spectrometry (MS) and genomes sequencing based PTM activity prediction, the number and types of protein PTMs detected and characterized in microbiota, mainly bacteria, have increased rapidly. Compared to eukaryotic proteins, most PTMs occur in a relatively small number of bacterial proteins with low levels of modification. Despite this, there is growing evidence that protein PTMs play an essential role in a variety of bacterial biological processes including protein synthesis and transformation, bacterial cell cycle, capsule and biofilm formation, stationary phase functions, persistence and virulence, and nitrogen metabolism. In addition, the number of modifying enzymes varies greatly among different species of bacteria, and the extent of the modified proteome is closely associated with environmental conditions. Depending on the protein and the conditions under which the host bacteria grow, the modified protein may behave differently.
| Known bacterial PTMs and their involvement in cellular physiology | |
|---|---|
| PTMs | Cellular processes |
| Phosphorylation | Regulation of adaptation, cellular metabolism, capsules and biofilm synthesis, protein quality control and recycling. |
| Acetylation | Regulation of enzymatic activities, cellular metabolism. |
| Glycosylation | Regulation of transmembrane and secreted proteins, cell-cell interaction, and virulence. |
| Pupylation | Regulation of protein quality control and recycling. |
| Lipidation | Control of protein anchoring and export. |
Creative Proteomics is a leading custom service provider in PTM proteomics analysis. With a team of experts and state-of-the-art equipment, our PTM proteomics analysis platform can help our customers to explore diverse protein PTMs in bacteria, promoting the understanding of bacterial physiology and opening new avenues for the treatment of infectious diseases. Our services adapt to the following lists, but are not limited to:
Our services are customer oriented and dedicated to providing a cost-effective and high-quality service experience. Please feel free to contact us for more details.
References
Our products and services are for research use only.