
- Home
- PTMs Proteomics
- Proteomics Analysis of ADP-Ribosylation
ADP-ribosylation is a pivotal post-translational modification process that plays a crucial role in cellular regulation and homeostasis. This alteration affects a number of biological processes by moving ADP-ribose moieties from NAD+ to substrate proteins. By exploring the complex field of ADP-ribosylation, Creative Proteomics seeks to clarify its significance, analytical techniques, uses, and crucial factors to take into account during the analytical procedure.
ADP-ribosyltransferases (ARTs) are a family of enzymes that transfer ADP-ribose units to target proteins, therefore catalyzing ADP-ribosylation. Numerous biological functions are impacted by this alteration, such as chromatin control, DNA repair, and cellular stress responses. ADP-ribosylation is a dynamic process with a wide range of substrates, making it an intriguing field of study with implications for both basic biology and medicinal development.

Analysis of ADP-ribosylation is a complex yet rewarding endeavor that sheds light on fundamental cellular processes and holds immense potential for therapeutic advancements. Creative Proteomics through leverage a combination of cutting-edge analytical methods can unravel the intricacies of ADP-ribosylation, paving the way for innovative solutions in cancer, neurodegenerative diseases, and drug development. As the field continues to evolve, a nuanced understanding of ADP-ribosylation dynamics will undoubtedly shape the future of biological research and clinical applications.
Profiling of the ADP-Ribosylome in Living Cells
Journal: Angew Chem Int Ed Engl
Published: 2022
Background
ADP-ribosylation modifications on proteins are related to DNA damage repair, transcriptional control, cell division, and other processes in living organisms, and in some cancers are treated with inhibitors of ADP-ribosylase. Linear or branching-type ADP-ribose undergoes a dynamic and reversible attachment of NAD+ as a substrate to a variety of amino acid residues of proteins Glu, Asp, Cys, Arg, Lys, Ser, and Tyr, which alters the electrical, structural, functional, and action network of proteins, but currently, there is a lack of favorable tools to study this modification for reasons that include the process of cellular fragmentation that can disrupt pre-existing ADP-ribosylation modifications, non-covalent ADP-ribose-binding proteins can segregate in a mixed fashion, and background ADP-ribosylation modifications can affect modifications that arise in response to external stimuli. NAD+ analogs have also been developed to aid in the study, but they have poor cell permeability, capture a limited number of proteins, and do not even identify known relevant enzymes, so further development is needed. In this paper, the authors have developed a new NAD+ analog probe that is very biocompatible and efficiently identifies protein changes in cells under a wide range of conditions.
Results
The authors first designed and synthesized 2 NAD+ analog probes with imaging motifs or desulfobiotin, respectively, which have been shown in the previous literature to have large motifs on the C2 of the molecule that can be accepted by ADP-ribosylase. Both probes synthesized by the authors were shown by on-gel imaging to label ADP-ribosylase, an enzyme that is capable of self-modification, and the labeling intensity diminished in response to the addition of natural NAD+. The authors then used both probes in cellular experiments, first using fluorescent NAD+ analogs to screen for optimal transfection reagent conditions that were both highly efficient in entering the cell and evenly distributed within the cell, and virtually non-toxic. Fluorescence microimaging showed that there was a clear fluorescent signal of the analogues in cells under the oxidative stress generated by hydrogen peroxide induction, while the signal disappeared with the addition of the enzyme inhibitor, suggesting that the fluorescent NAD+ analogues synthesized by the authors metabolize the labeled ADP-ribosylase well in cells (Figure 1).
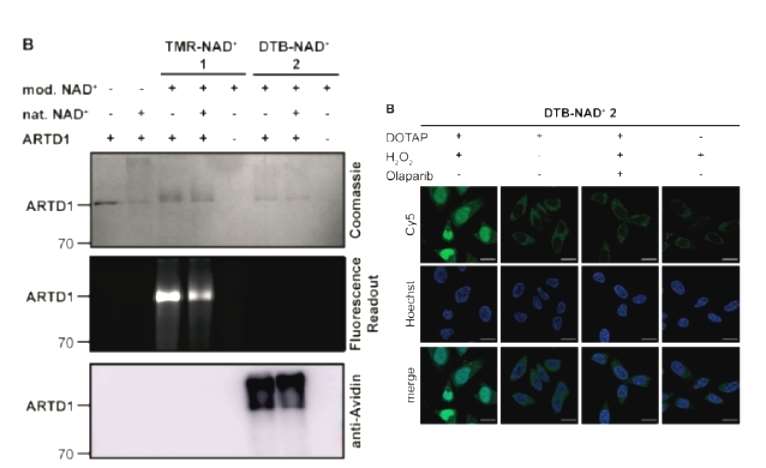 Figure 1
Figure 1
Next, for the desulfobiotin NAD+ analog, the authors demonstrated that several other members of the ADP-ribosylation enzyme family were well labeled, and so, after testing the metabolic labeling ability of this desulfobiotin analog in cells by borrowing the transfection conditions of the fluorescent analog, the authors designed a proteomics process in hopes of identifying the ADP-ribosylation-related proteins. Firstly, the analog was transferred into cells, and a blank group, an H2O2-treated group, and an H2O2+ADP-ribosylase inhibitor-treated live cell group was set up, and then the cells were lysed, enriched to obtain modified proteins, washed and eluted, and then these proteins were removed from modification again, enzymatically digested, and then identified by mass spectrometry, which identified 310 more plausible targets from 2067 proteins after rigorous screening. The authors concluded that the methodological process was stable and reliable and was able to identify both known and unknown ADP-ribosylation-associated proteins (Figure 2).
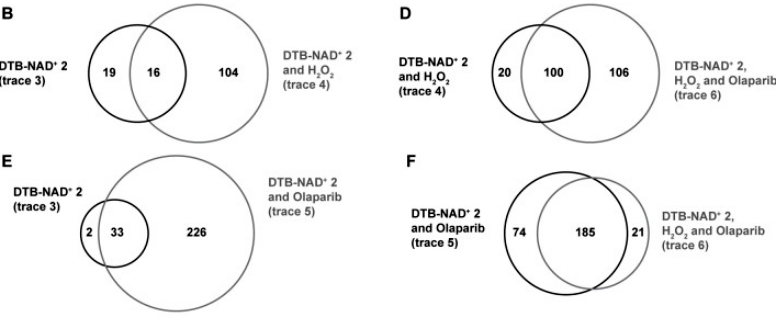 Figure 2
Figure 2
Conclusion
This study demonstrates that the utility of the newly developed NAD+ analogs in combination with our mild and effective cell delivery protocol enables imaging of PAR in living cells and to analyze the cellular adaptation by protein ADP‐ribosylation as a consequence of environmental changes such as H2O2‐induced oxidative stress or the effect of drugs such as olaparib. These findings are therefore paving the way for further functional and clinical studies of the ADP‐ribosylated proteome in living cells in health and disease.
Reference
Our products and services are for research use only.