
- Home
- DNA/RNA Modification
- mRNA Modification LC-MS Analysis Service
Messenger RNA (mRNA) plays a crucial role in regulating cellular functions by directly translating proteins. Various modifications can occur on mRNA, distributed widely across the 5' cap, 5' UTR, coding sequence (CDS), or 3' UTR regions. Specifically, the 5' cap structure contains multiple modifications, including m7G, 2'-O-methyl (Nm), and N6,2'-O-dimethyladenosine (m6am). Among these modifications, m6am is believed to be exclusively present in the 5' cap region, while Nm and m7G may also be found in the transcriptome.
N6-methyladenosine (m6A) is the most abundant modification on mRNA, with an average of 1 to 3 m6A modifications per transcript. It is typically highly enriched near the stop codon region. mRNA m6A modification is the most extensively studied and researched modification type. It is added by the modification enzyme complex composed of METTL3 and METTL14, and removed by the demethylase complex consisting of FTO/ALKBH5. The m6A modification affects mRNA's alternative splicing, nuclear-cytoplasmic transport, stability, and translation efficiency through interactions with various m6A reader proteins. Defects and alterations in mRNA m6A modification and modifying enzymes are closely associated with human diseases, such as cancer, neurodevelopmental and neurological disorders, cardiovascular diseases, immunity and inflammation, stem cell differentiation, reproduction, metabolism, and obesity. Furthermore, other modifications, such as m5C added by NSUN2/DNMT2/TRDMT1, can impact mRNA's alternative splicing, nuclear-cytoplasmic transport, stability, and translation efficiency and are associated with cancer, early embryonic development, and viral infections.
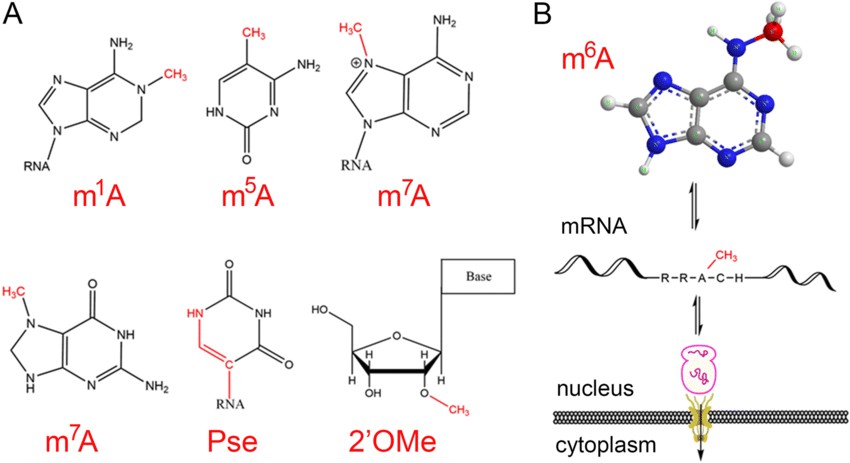 Figure 1 Different types of mRNA modifications
Figure 1 Different types of mRNA modifications
To detect the changes in the abundance of different modifications on mRNA, Creative Proteomics provides LC-MS analysis services for mRNA modification quantification. This service involves the extraction of total RNA from cells or tissues, isolation of mRNA, complete hydrolysis and dephosphorylation to generate individual modified and unmodified nucleosides. The LC-MS system is then used to quantitatively analyze and compare the abundance and proportions of each modification or unmodified nucleoside in the experimental and control groups.
Specifically, nucleosides with different masses and polarities have different retention times in the HPLC high-performance liquid chromatography, allowing for the preliminary separation of different modified nucleosides. Subsequently, these nucleosides undergo ionization and enter the mass spectrometer. Based on the characteristic parent ion-to-fragment ion mass-to-charge ratio of each nucleoside and the retention time in HPLC, peak areas are extracted for quantitative analysis of modified nucleosides.
Optimized pre-treatment and the use of high-quality Agilent equipment ensure comprehensive and accurate analysis of base modifications, elevating the sensitivity, precision, dynamic range, and robustness of the analysis results to a new level.
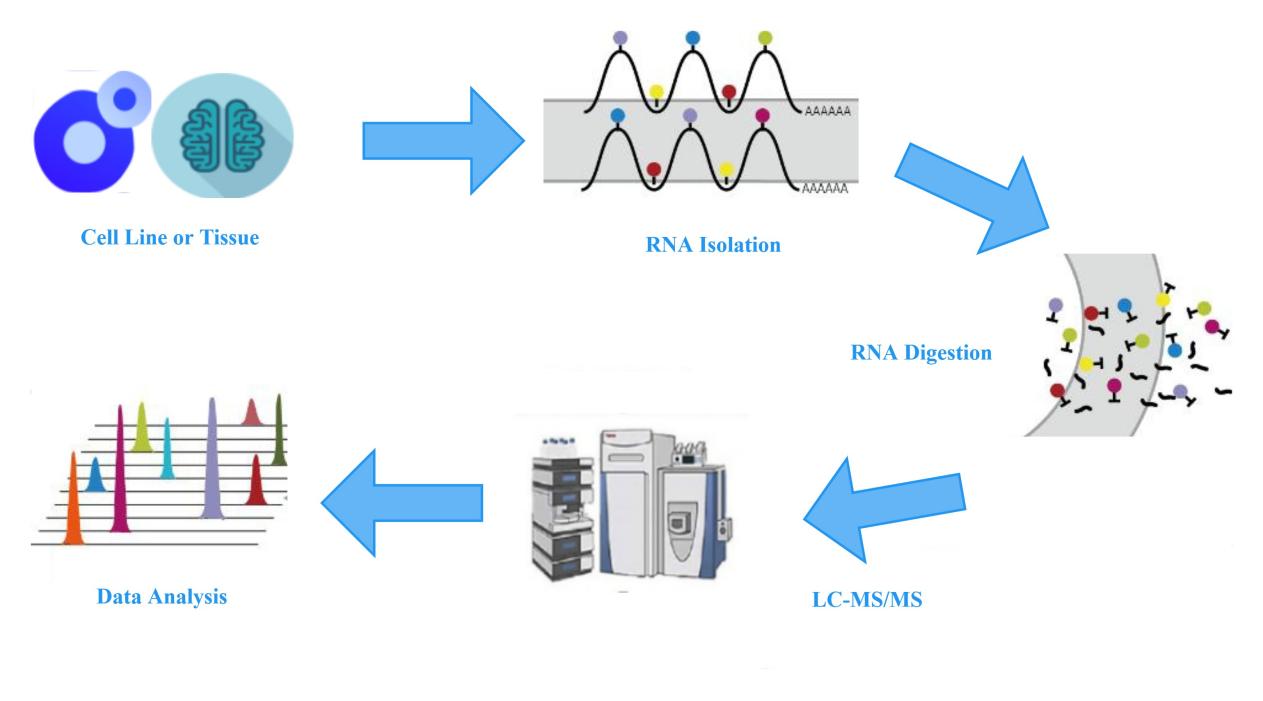 Figure 2: Experimental workflow for LC-MS measurement of mRNA modifications.
Figure 2: Experimental workflow for LC-MS measurement of mRNA modifications.
| Number | Nucleoside | Symbol | Number | Nucleoside | Symbol |
|---|---|---|---|---|---|
| 1 | 3′-O-methyladenosine | 3′-OMeA | 27 | 3'-O-methyluridine | 3'-OMeU |
| 2 | 2′-O-methylcytidine | Cm | 28 | 5-methyl-2-thiouridine | m5s2U |
| 3 | 3-methylcytidine | m3C | 29 | 5-methoxyuridine | mo5U |
| 4 | 5-methylcytidine | m5C | 30 | pseudouridine | Ψ |
| 5 | N6-isopentenyladenosine | i6A | 31 | 2'-O-methylinosine | Im |
| 6 | 5,2'-O-dimethylcytidine | m5Cm | 32 | 3-methyluridine | m3U |
| 7 | 1-methyladenosine | m1A | 33 | 1-methylpseudouridine | m1Ψ |
| 8 | 2-thiocytidine | s2C | 34 | 5-hydroxymethylcytidine | hm5C |
| 9 | N2,N2,7-trimethylguanosine | m2,2,7G | 35 | 5,2'-O-dimethyluridine | m5Um |
| 10 | N4-acetyl-2'-O-methylcytidine | ac4Cm | 36 | N6-threonylcarbamoyladenosine | t6a |
| 11 | N6-methyladenosine | m6A | 37 | 2-methylthio-N6-threonylcarbamoyladenosine | ms2t6A |
| 12 | 3'-O-methylcytidine | 3′-OMeC | 38 | 5-carboxymethyluridine | cm5U |
| 13 | 2'-O-methyladenosine | Am | 39 | 5-methoxycarbonylmethyl-2-thiouridine | mcm5s2U |
| 14 | N2, N2-dimethylguanosine | m22G | 40 | 5-Methoxycarbonylmethyluridine | mcm5U |
| 15 | 5'-O-methylthymidine | 5′-OMeT | 41 | 2-methylthio-N6-isopentenyladenosine | ms2i6A |
| 16 | 2′-O-methyluridine | Um | 42 | Peroxywybutosine | o2Yw |
| 17 | inosine | I | 43 | 5-taurinomethyl-2-thiouridine | tm5s2U |
| 18 | 2′-O-methylguanosine | Gm | 44 | 5-oxyacetic acid uridine | cmo5U |
| 19 | 1-methylguanosine | m1G | 45 | 5-carbamoylmethyuridine | ncm5U |
| 20 | 7-methylguanosine | m7G | 46 | Queuosine | Q |
| 21 | N2-methylguanosin | m2G | 47 | 5-taurinomethyluridine | tm5U |
| 22 | 3'-O-methylinosine | 3'-OMeI | 48 | 5-formyl-2′-O-methylcytidine | f5Cm |
| 23 | 2-thiouridine | s2U | 49 | dihydrouridine | D |
| 24 | 4-thiouridine | s4U | 50 | 5-formylcytidine | f5c |
| 25 | 5-methyluridine | m5U | 51 | wybutosine | yW |
| 26 | N4-acetylcytidine | ac4C | 52 | 5-methoxycarbonylmethyl-2'-o-methyluridine | mcm5Um |
Our products and services are for research use only.