
- Home
- PTMs Proteomics
- Proteomics Analysis of Pupylation
Pupylation, akin to ubiquitination in eukaryotes, involves the conjugation of Pup to specific lysine residues on target proteins. This modification is catalyzed by a cascade of enzymes, including the Pup ligase (PafA) and deamidase (Dop). The tagging of proteins with Pup serves as a signal for recognition by the mycobacterial proteasome, leading to targeted degradation. This finely tuned regulatory mechanism plays a pivotal role in maintaining cellular homeostasis.
Pupylation stands as a captivating area of research, offering profound insights into bacterial physiology and cellular regulation. Creative Proteomics follows advanced analytical methods and technologies that continue to propel pupylation analysis into the forefront of proteomics research, and we provide pupylation analysis that not only deepens our understanding of bacterial biology but also offers great potential for therapeutic and diagnostic advances in the field of infectious diseases.
Pupylation of PafA or Pup inhibits components of the Pup-Proteasome System
Journal: FEBS Lett
Published: 2018
Background
Ubiquitin (Ub) is attached to lysine residues on target proteins, forming polyubiquitin (polyUb) chains with different lysine linkages. While K48-linked chains are associated with proteasomal degradation, other linkages, like K63, have nondegradative roles. The article highlights a Ub-like protein, Pup, found in Actinobacteria and some bacterial lineages. Pup is a small, natively unstructured protein conjugated to lysine residues by the Pup ligase, PafA. The attached proteins are recognized by the AAA+ protein Mpa and the mycobacterial 20S proteasome for ATP-dependent turnover. Pup, like Ub, has conserved lysine residues, and the article proposes that polyPup chains could form with different biological activities. Recent research by Gur and colleagues showed in vitro polypupylation of Pup primarily through K61. The article supports these findings, demonstrating that polypupylation of Pup and model substrates occurs preferentially through K61. It further reveals that polyPup, whether free or attached to a target protein, can inhibit proteasome function.
Results
The article explores the role of lysine residues (K7, K31, and K61) in polyPup chain formation in MsmPup. Through mutagenesis and cloning, single, double, and triple point mutants of Pup(E) were generated. These mutants were examined for their ability to conjugate to model target proteins, PanB and E. coli ClpS. All mutants efficiently conjugated to PanB, indicating no compromise in Pup(E) structure or function. Examining self-pupylation, K7 or K31 mutations had minimal effects, while K61 mutation significantly reduced polyPup formation by approximately 80%. Combining K61 mutation with K7 or K31 mutations abolished polyPup formation. Importantly, all mutant Pup forms could pupylate the model substrate (EcClpS), similar to wild-type Pup. The study suggests that polyPup formation predominantly occurs through the conserved K61, raising speculation about its potential regulatory or nonproteolytic role in the cell (Figure 1).
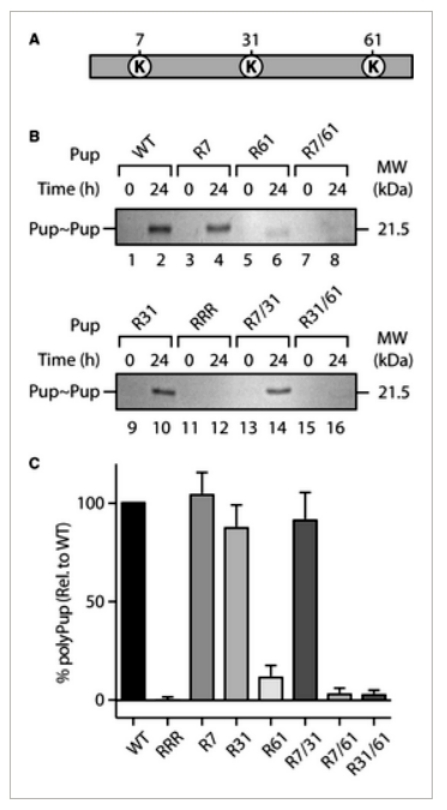 Figure 1
Figure 1
The study investigates the impact of various Pup mutants on Mpa-mediated turnover of pupylated proteins by the 20SOG proteasome. Mutants were generated through quick change mutagenesis, and their ability to conjugate to model target proteins, PanB and E. coli ClpS, was assessed. The rate of turnover of monopupylated PanB was found to be largely independent of the specific Pup mutant conjugated. However, when examining the turnover of a model protein, GFP, different results were observed. Multipupylated GFP was rapidly degraded, while GFP containing a mixture of poly- and multipupylation forms showed little to no turnover, suggesting that polyPup inhibits substrate turnover.
Further experiments demonstrated that free polyPup inhibited the proteasome-mediated turnover of multipupylated GFP and monopupylated PanB. This inhibition was specific to polyPup, as free Pup had no effect. Investigating the proteasome components affected by polyPup, it was found that polyPup did not impact the turnover of a fluorescently labeled peptide substrate (LLVY-amc) by the 20SOG CP. However, polyPup stimulated the ATPase activity of Mpa similarly to a model monopupylated substrate (PanB~Pup). The study proposes that polyPup interacts with the coiled-coil domain of Mpa, cross-linking multiple domains and blocking substrate translocation through the Mpa pore. This mechanism protects polypupylated substrates, including polyPup itself, from proteasomal degradation. The findings suggest a potential role for polyPup in reversibly regulating proteasome activity and nonproteolytic regulation of specific target proteins. The study acknowledges the existing challenge in determining the rules governing target protein pupylation. Nonetheless, the discovery that PafA, the Pup ligase, can be multipupylated adds to the complexity of this regulatory system (Figure 2).
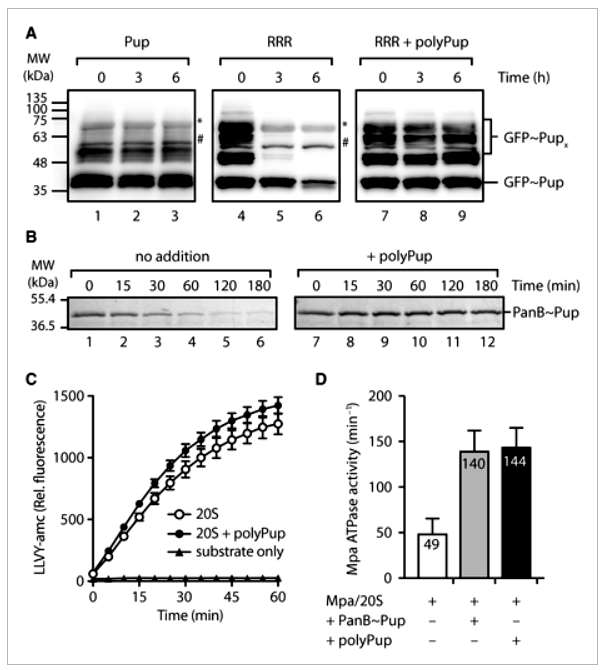 Figure 2
Figure 2
The research investigates single- and double-lysine mutants of Pup, revealing a significant increase in high molecular weight pupylation of PafA, likely representing polypupylation. Unlike polyPup formation, polypupylation of PafA appears to occur through all three lysine residues (K7, K31, and K61) in Pup. The reasons for this discrepancy remain unclear. The significance of PafA oligopupylation (a mixture of polypupylation and multipupylation) is explored by monitoring PafA enzyme activity before and after autopupylation. Pup conjugation to PanB is significantly reduced following autopupylation, indicating that oligopupylated PafA inhibits its activity. This inhibition is observed with both wild-type Pup and the RRR mutant, suggesting that multipupylation alone is sufficient to inhibit PafA enzyme activity. These findings parallel the regulatory pupylation observed in Mpa, emphasizing the intricate control of the Prokaryotic Proteasome System (PPS) (Figure 3).
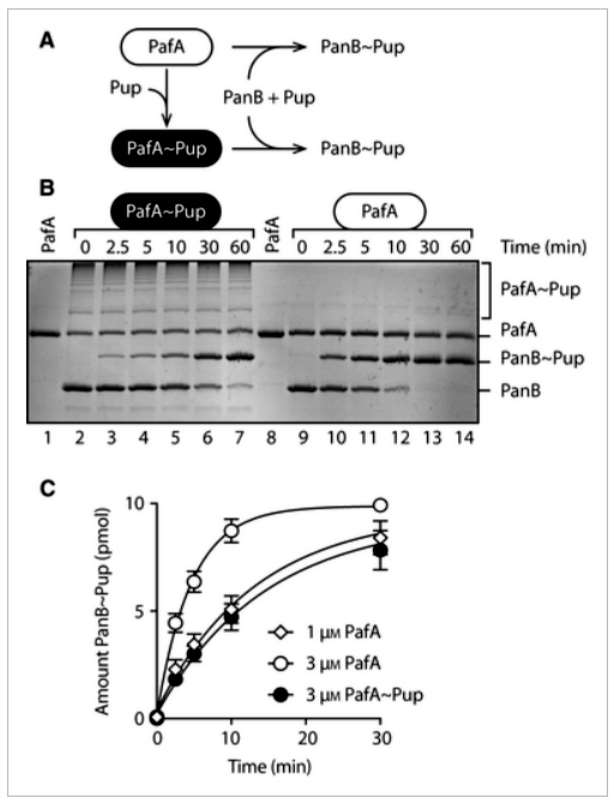 Figure 3
Figure 3
Conclusion
The study reveals that polypupylation of Pup, particularly through lysine residue K61, aligns with recent findings by Gur and colleagues. It is demonstrated that polyPup, whether free or attached to a target protein, inhibits proteasome function. The proposed mechanism involves the cross-linking of the coiled-coil (CC) domains of Mpa, hindering substrate translocation through the Mpa pore and preventing entry into the 20S CP, thereby blocking substrate turnover.
Additionally, the study explores PafA autopupylation, which occurs at multiple lysine residues. Optimal autopupylation involves three lysine residues, likely leading to polypupylation. In contrast to Pup, the polypupylation of PafA is suggested to be heterotypic, as single or double lysine mutants of Pup do not prevent the formation of high molecular weight PafA conjugates. Autopupylation of PafA, occurring in the absence of an additional substrate, completely inhibits the ligase activity of PafA. The authors propose that PafA autopupylation serves as a regulatory mechanism to reversibly control target protein pupylation. In the absence of a target protein, autopupylation inhibits PafA ligase activity; however, when target proteins are abundant, depupylation of PafA~Pup, likely mediated by Dop, results in the production of free Pup for conjugation and an active enzyme for pupylation. The in vitro data collectively suggest that pupylation, whether of Pup (polyPup) or PafA, may regulate different components of the Pup-Proteasome system (PPS).
Reference
Our products and services are for research use only.