
- Home
- PTMs Proteomics
- Proteomics Analysis of Adenylylation
Adenylylation, a pivotal biochemical process, involves the covalent attachment of adenosine monophosphate (AMP) to various biomolecules, playing a crucial role in cellular signaling, regulation, and homeostasis. This enzymatic modification, catalyzed by adenylyl transferases, has far-reaching implications in diverse biological contexts.
Adenylylation often interplays with other post-translational modifications (PTMs), creating a complex network of regulatory events. Investigating the cross-talk between adenylylation and acetylation, phosphorylation, or ubiquitination expands our comprehension of cellular regulatory networks. Dysregulation of adenylylation is implicated in various diseases, including cancer and neurodegenerative disorders. Unraveling the molecular basis of aberrant adenylylation opens avenues for therapeutic interventions targeting these diseases.
Analysis of adenylylation is important in many biological and biochemical applications. Adenosine monophosphate (AMP) moiety covalently attaches itself to a target molecule, which is typically a protein or nucleic acid, in this process. As a post-translational alteration that can control the activity, location, and interactions of biomolecules, this mechanism is essential to many different cellular processes.
Our researchers can assist other researchers in gaining understanding of the complex molecular pathways underpinning cellular functions by doing adenylylation analysis. Gaining knowledge about the precise locations and circumstances of adenylylation facilitates a more profound understanding of the role these alterations play in the general control of biological processes. This information is essential for improving our comprehension of gene expression, cellular signaling, and the molecular causes of numerous illnesses.
Adenylylation stands as a cornerstone in the intricate web of cellular regulation. From its molecular intricacies to its far-reaching implications in health and disease, this post-translational modification beckons further exploration. The integration of advanced analytical techniques, coupled with a comprehensive understanding of its applications, propels adenylylation research into the forefront of modern molecular biology. Creative Proteomics relies on advanced analysis techniques, continue to delve deeper of adenylylation, the prospect of harnessing the therapeutic potential of adenylylation modulation emerges, promising a paradigm shift in precision medicine.
Chemoproteomic discovery of a human RNA ligase
Journal: Nat Commun
Published: 2023
Background
RNA ligases play an important role in RNA sealing and are mainly involved in biogenesis processes such as intron-containing tRNA splicing, tRNA repair, mRNA splicing in the unfolded protein reaction (UPR), and RNA recombination. RNA ligases are known to link the 5'-PO4 and 3'-OH ends of RNAs by a classical three-step mechanism: first, the RNA ligase undergoes 5'-adenosine triphosphate (ATP)-dependent auto-AMPylation (also known as auto-adenylation) at the catalytic lysine group. Subsequently, AMP is transferred from ligase-(lysine-N)-AMP to the 5'-PO4 end of the RNA (pRNA) to produce an RNA-adenylation intermediate (AppRNA). Finally, upon nucleophilic attack of the 3'-OH of the AppRNA, the two RNA ends are joined by a phosphodiester bond, releasing AMP.
This type of enzyme, known as 5'-3' RNA ligase, is widely used in RNA editing and sequencing, but this enzyme remains to be characterized in vertebrates, and only one protein RNA ligase has been identified in humans, a guanidinium 5'-triphosphate (GTP)-dependent 3 '-5' RNA ligase, which connects the 2', 3'-cyclic phosphate (cPO4) and 5'-OH ends of RNA as a subunit of the human tRNA splice ligase complex. Therefore, in this paper, the authors synthesized a chemical probe based on adenosine triphosphate (Ap3A) for the discovery of a human 5'-3' RNA ligase.
Results
AMPylation is a PTM in which an AMP molecule covalently binds to the side chain or C-terminus of a protein. First the authors synthesized a C2,C2′-ethynyl-modified Ap3A (C2-eAp3A) probe that mimics ATP and is stable in cell lysates. Using this probe, the authors performed an activity-based proteomics analysis (ABPP) to enrich for proteins capable of undergoing AMPylation modifications. Based on this approach, they identified an unknown functional protein from the open reading frame of chromosome 12, C12orf29. To confirm that it undergoes AMP modification, the authors constructed recombinant wild-type C12orf29 and expressed it in E. coli. They found that the purified protein had a molecular weight 329 Da higher than the theoretical molecular weight, which is consistent with the molecular weight of the AMPylated modification. In addition, structure prediction by AlphaFold showed that the authors found that the protein had similar structural domains to NgrRnl, an RNA ligase identified from Griseofulvin (Figure 1).
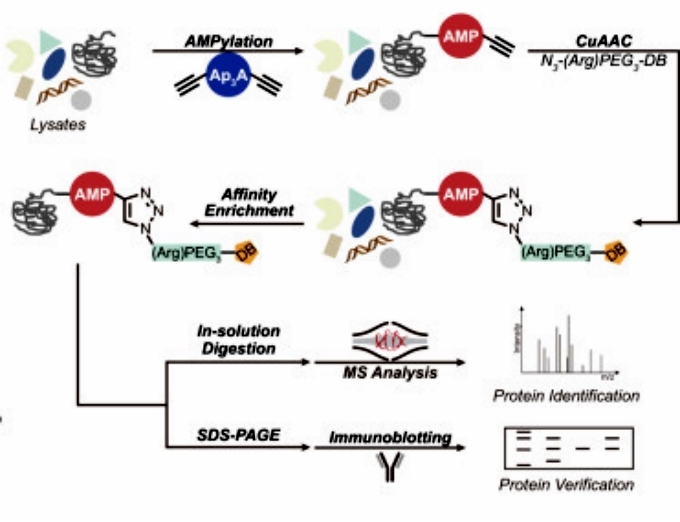 Figure 1
Figure 1
In further studies, the authors found that C12orf29 was able to form C12orf29-AMP using ATP as a substrate in the presence of Mg2+ as a cofactor. and they found that non-phosphorylated 3'-RNA ends were essential for C3orf12-mediated RNA ligation using single-stranded DNA (ssDNA) as a substrate. Subsequently, to investigate the biological function of C12orf29, the authors constructed C12orf29-KO HEK293 cells. WT and KO cells were treated with menaquinone, a cellular stress molecule known to produce ROS, respectively, and the authors found that the cell viability of WT was significantly higher than that of the KO group, and that 28SRNA was more susceptible to degradation in the KO group, suggesting that C12orf29 plays an important role in the maintenance of RNA integrity as well as cellular resistance to ROS damage (Figure 2).
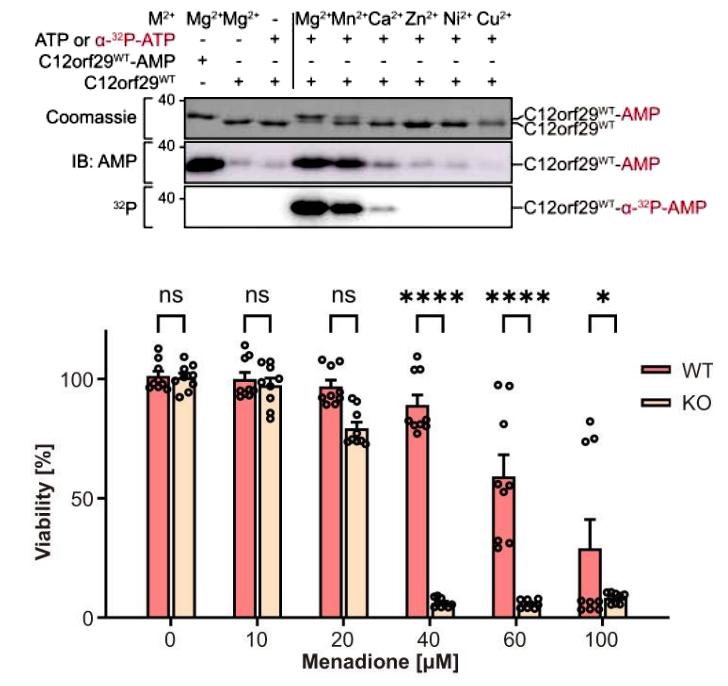 Figure 2
Figure 2
Conclusion
In this paper, the authors designed an Ap3A-based chemical probe to interrogate the human AMPylated proteome. As a structural analog of ATP, Ap3A shows superior chemical and enzymatic stability under reaction conditions, making the Ap3A-based probe promising for enriching AMPylated proteins from cell lysates without the need for external AMPylators. Using the Ap3A-based chemical probe, the authors identified C12orf29 as a 5'-3' RNA ligase, which has been demonstrated in detail in this paper by a three-step catalytic mechanism of ligating 5'-PO4 within the single-stranded region through and RNA-AMPylation of the 3'-OHRNA ends, designed an Ap3A-based chemical probe that can characterize the human AMPylated proteome, and utilized this approach to find a new RNA ligase, C12orf29.
Reference
Our products and services are for research use only.