
- Home
- PTMs Proteomics
- Post-Translational Modification of Protein Biopharmaceuticals
- Yeast Glycosylation Analysis
Yeast glycosylation, a pivotal post-translational modification (PTM), plays a crucial role in the structural and functional diversity of proteins. This intricate process involves the enzymatic addition of glycan moieties to proteins, influencing their stability, localization, and biological activity. Understanding yeast glycosylation is fundamental for deciphering the molecular intricacies governing cellular processes. Yeast glycosylation as a prevalent PTM, significantly impacts the molecular characteristics of proteins, leading to a spectrum of functional outcomes.
Yeast, particularly Saccharomyces cerevisiae, serves as an exemplary model organism for studying glycosylation due to its well-defined genetics and eukaryotic cellular processes. The glycosylation pathways in yeast involve the Endoplasmic Reticulum (ER) and Golgi apparatus, where sequential enzymatic reactions culminate in the addition of oligosaccharide chains to specific protein residues.
Yeast glycosylation analysis stands at the forefront of molecular research, unraveling the intricacies of protein modifications with profound implications in biotechnology, medicine, and basic science. As technology continues to evolve, so too will our ability to decode the language of glycosylation, providing unprecedented insights into cellular processes and opening new avenues for therapeutic interventions. Creative Proteomics has rich service experience in Yeast Glycosylation Analysis, if you are interested in us, please feel free to contact us.
Quantitative Profiling of N-linked Glycosylation Machinery in Yeast Saccharomyces cerevisiae
Journal: Mol Cell Proteomics
Published: 2018
Background
Asparagine-linked glycosylation is a common posttranslational protein modification regulating the structure, stability and function of many proteins. The N-linked glycosylation machinery involves enzymes responsible for the assembly of the lipid-linked oligosaccharide (LLO), which is then transferred to the asparagine residues on the polypeptides by the enzyme oligosaccharyltransferase (OST). A major goal in the study of protein glycosylation is to establish quantitative methods for the analysis of site-specific extent of glycosylation. We developed a sensitive approach to examine glycosylation site occupancy in Saccharomyces cerevisiae by coupling stable isotope labeling (SILAC) approach to parallel reaction monitoring (PRM) mass spectrometry (MS). We combined the method with genetic tools and validated the approach with the identification of novel glycosylation sites dependent on the Ost3p and Ost6p regulatory subunits of OST.
Results
The author focuses on developing a targeted mass spectrometry (MS) approach for quantitatively profiling N-linked glycoproteins in yeast using Stable Isotope Labeling by Amino acids in Cell culture (SILAC) combined with Parallel Reaction Monitoring (PRM). The assay development involves enriching glycopeptides from microsomal fractions through proteinase digestion, with the use of LysC and trypsin for increased yield and accuracy. Zwitterionic hydrophilic interaction chromatography (ZIC-HILIC) is employed for solid-phase extraction (SPE) to enhance the coverage of hydrophilic N-linked glycopeptides. The glycans are released using endoglycosidase H, providing an advantage over other methods using PNGase F for deglycosylation. The study successfully identifies 166 high-confidence glycosylation sites corresponding to 105 glycoproteins, with the majority represented by one or two glycopeptides. This data forms the basis for a PRM-based approach, where glycopeptides are monitored alongside non-glycosylated peptides from the same glycoprotein (Figure 1).
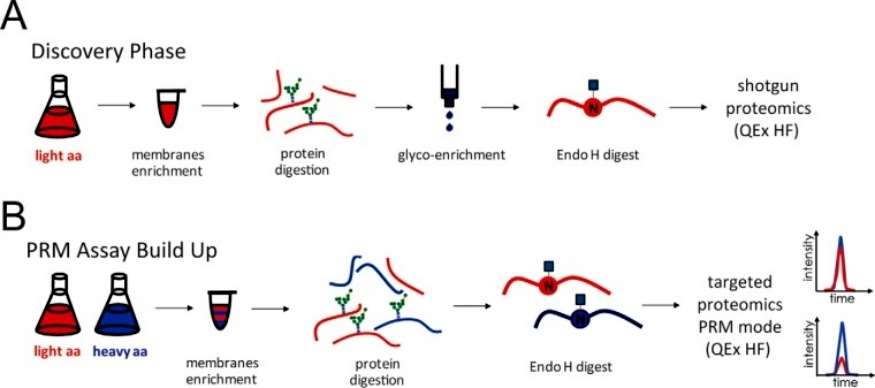 Figure 1
Figure 1
The SILAC PRM MS analysis revealed that in the yeast strain lacking the Ost3 protein, 84% of glycosylation sites were hypoglycosylated (occupancy ratio < 0.75 compared to the wild-type reference strain). In contrast, the strain overexpressing OST6 showed less severe hypoglycosylation, with 39% of glycosylation sites being affected. The severe hypoglycosylation in the Δost3 strain was attributed to both a reduced level of fully assembled OST and the absence of OST3 function. These findings underscore the importance of using yeast strains expressing normal levels of fully assembled OST but lacking specific subunits for functional analysis. In strains lacking Ost6p, only 2% of glycosylation sites were hypoglycosylated when OST3 was overexpressed, indicating that the Ost3p-containing complex has broader substrate specificity compared to the Ost6p-containing complex. This aligns with previous research suggesting a more significant role for Ost3p in N-linked glycosylation. Ost3p affects polypeptide substrate specificity, as evidenced by hypoglycosylation on a subset of glycosylation sites in strains with Ost6p-containing OST. The study identified 17 novel Ost3p substrates, with glycosylation dependence on the presence of the Ost3 subunit. Notably, these substrates, when structurally examined, mostly required oxidative folding (disulfide bond formation), supporting the proposed function of Ost3 protein in transiently forming mixed disulfide bonds with OST polypeptide substrates (Figure 2).
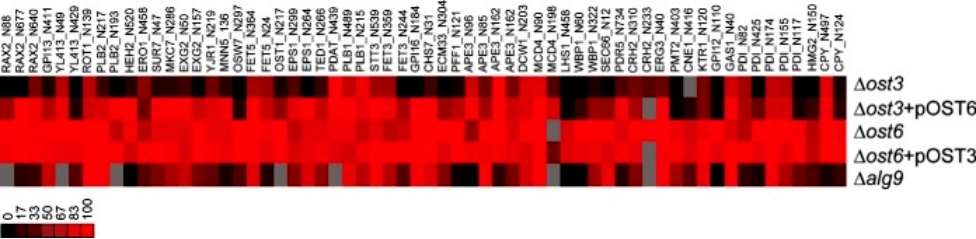 Figure 2
Figure 2
Sequence analysis was conducted on glycosylation sequons along with ten residues upstream and downstream from the glycosylation sites. Two-sample logo analysis was utilized to visualize differences between efficiently and poorly glycosylated sequences around these sites. For glycosylation sites influenced by Ost6-containing OST (in Δost3 strain complemented with OST6), the efficiency was not dependent on the presence of specific amino acids in the local environment of the glycosylation site. In contrast, poorly glycosylated sites in the Δalg9 strain were enriched in N-X-S sequons, as opposed to the more efficiently modified N-X-T sequons. Sequons containing glutamic acid at the +1 position were also less likely to be efficiently glycosylated. Additionally, sites surrounded by basic arginine and lysine residues, as well as polar amino acids, were less favored when presented with a suboptimal glycan donor. Examining the localization of efficiently glycosylated sites in relation to their structural environment, for substrates with available structure or structural models, revealed that glycosylation efficiency was not dependent on the type of secondary structures on which glycosylation sites were located in Δost3 strains or Δalg9 strain. (Figure 3).
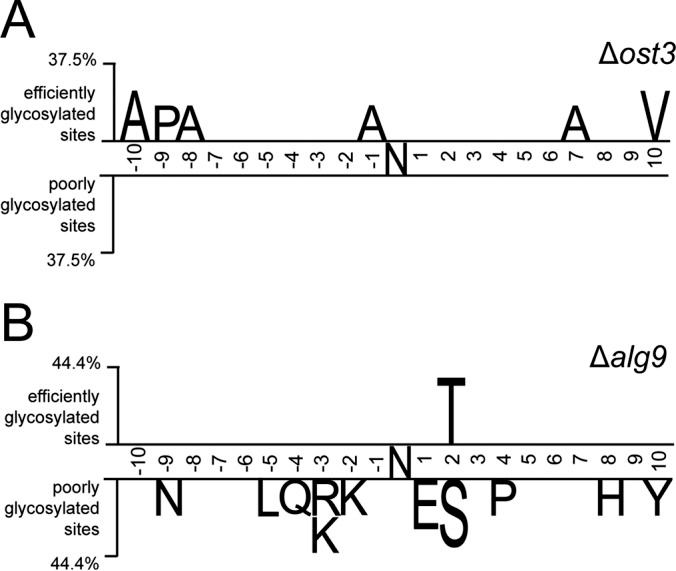 Figure 3
Figure 3
The author employed a novel analytical approach to delve into the N-glycosylation process of specific proteins, with a focus on the ER resident protein disulfide isomerase, Pdi1p, which carries five N-linked glycans. Pdi1p is structured with four thioredoxin folds (a, b, b', and a') that can fold independently. The a and a' domains each contain one disulfide bond directly involved in the catalytic activity of the isomerase. The five glycosylation sites are strategically located in structured areas of the protein: site 1 (N82, domain a) and site 2 (N117, domain a) are in structured loops, while site 3 (N155, domain b), site 4 (N174, domain b), and site 5 (N425, domain a') are situated in α-helices (Figure 4).
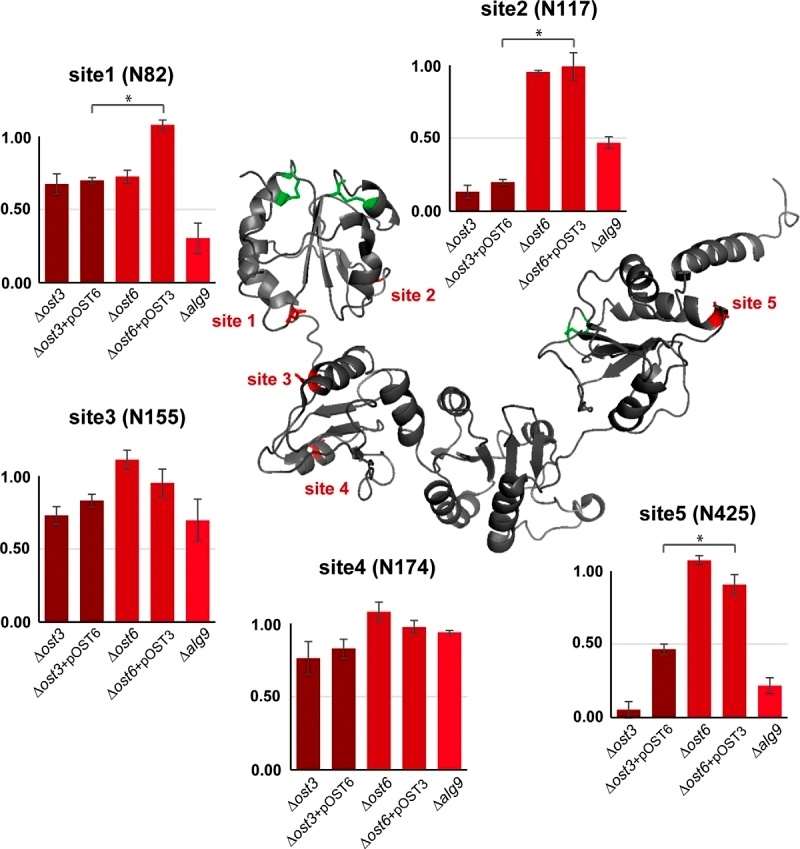 Figure 4
Figure 4
The author investigated the impact of defects in lipid-linked oligosaccharide (LLO) biosynthesis on the ergosterol pathway. Relative protein abundance of glycoproteins was determined across different strains using Parallel Reaction Monitoring (PRM) mass spectrometry, focusing on peptides without glycosylation sequons from 42 glycoproteins. A correlation was observed between hypoglycosylation and the unfolded protein response (UPR). Specifically, Δost3 and Δalg9 strains exhibited a significant increase in six proteins regulated by the UPR, including Ero1p, Rot1p, Lhs1p, Pdi1p, Mcd4p, and Eps1p. These proteins typically function as molecular chaperones and are involved in endoplasmic reticulum-associated protein degradation (ERAD) or UPR processes. Notably, ERG3 C-5 sterol desaturase, a protein in the ergosterol biosynthesis pathway, showed a four-fold increase in abundance in the Δalg9 strain but not in strains with mutations affecting OST components (Figure 5).
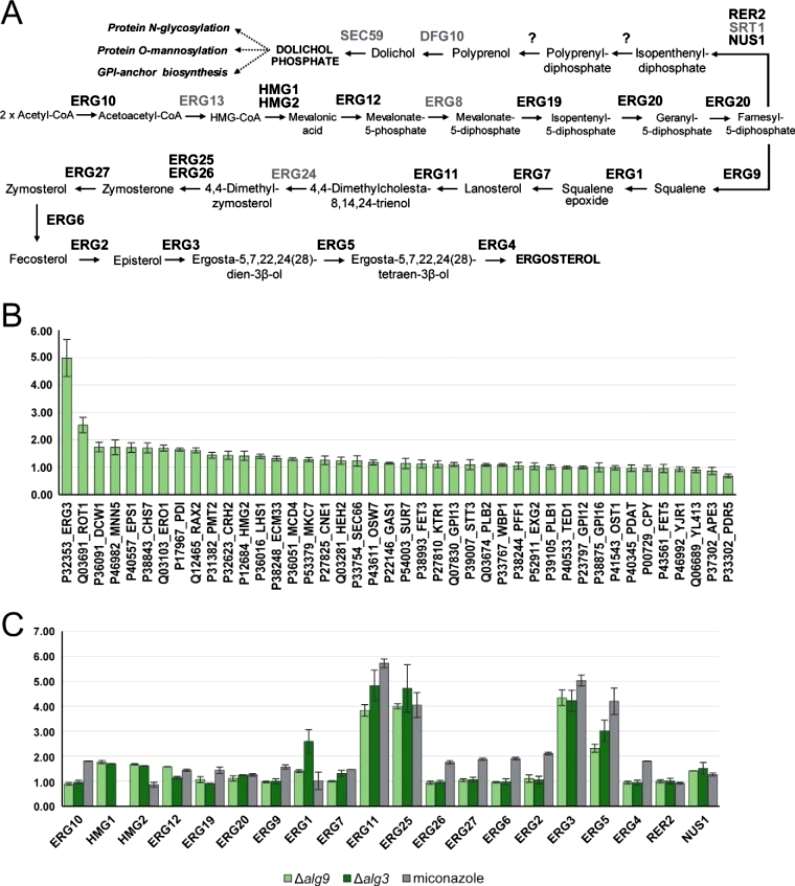 Figure 5
Figure 5
Conclusion
The researchers developed a sensitive method for assessing glycosylation site occupancy in Saccharomyces cerevisiae. This involved combining stable isotope labeling (SILAC) with parallel reaction monitoring (PRM) mass spectrometry (MS), and the method was further enhanced with genetic tools. Validation of the approach was achieved by identifying new glycosylation sites that rely on the Ost3p and Ost6p regulatory subunits of OST. The study concludes that the catalytic subunit Stt3p is directly involved in sequon recognition, while auxiliary subunits like Ost3p and Ost6p expand the substrate range of OST by influencing pathways such as protein folding. Notably, the research uncovered a novel regulatory network connecting isoprenoid lipid biosynthesis and lipid-linked oligosaccharide (LLO) substrate assembly.
Reference
Our products and services are for research use only.