
- Home
- PTMs Proteomics
- Proteomics Analysis of Nitration
In the field of biological research, nitration—a chemical process that involves adding a nitro group (-NO2) to a molecule—is essential. Because of this method's many applications and ramifications across different scientific areas, it has attracted a lot of interest. Fundamentally, nitration is the process of replacing a hydrogen atom with a nitro group. Reactive nitrogen species, frequently in the presence of catalysts or in particular environmental circumstances, accelerate this chemical transition. Researchers to utilize the nitration's potential applications must comprehend the mechanistic details of this process.

Creative Proteomics has always been at the forefront of scientific discovery, and based on our state-of-the-art technology platforms and specialized research services, we provide researchers with a multifaceted perspective on the intricate chemical modifications in biomolecules. From analytical methods that reveal molecular structure to applications spanning pharmaceuticals and environmental monitoring, our nitration research services are continually being used to unravel new chapters in the intricate symphony of life's molecules.
Protein Tyrosine Nitration in Plant Nitric Oxide Signaling
Journal: Front Plant Sci
Published: 2022
Background
This article delves into the intricate interplay between nitric oxide (NO) signaling and downstream events in plant physiology, particularly focusing on the role of transcription factors, such as ERFVIIs, in regulating various processes, including responses to hypoxia and ABA-mediated seedling establishment. The involvement of post-translational modifications (PTMs), especially protein nitration, is highlighted, as NO's reactivity with other molecules results in modifications like S-nitrosylation and tyrosine nitration. The study emphasizes the need to unravel the regulatory mechanisms underlying these PTMs and their impact on diverse physiological processes in plants.
Technical Approach
The technical approach involves employing mass spectrometry techniques, particularly liquid chromatography followed by tandem mass spectrometry (LC–MS/MS), to identify and analyze nitration sites in proteins. The study also utilizes immunoprecipitation (IP) with anti-3-nitroTyr antibodies for the enrichment of nitrated proteins, addressing the challenge posed by their low abundance and inherent instability. Additionally, the text discusses genetic code expansion technologies as a promising methodology for studying protein nitration. The approach combines in vivo and in vitro strategies, including exogenous treatment with nitrating agents and genetic strategies using mutant plants with increased peroxynitrite content to enhance the levels of detected nitrated proteins.
Results
Protein that are targets of NO-related PTMs can be simultaneously or sequentially modified by other PTMs, which can modify their functions by triggering conformational changes, hampering ligand or cofactor binding, translocating them to different subcellular localization, promoting their preteasome-mediated degradation, or even altering the functional interaction with other signaling pathways (Figure 1).
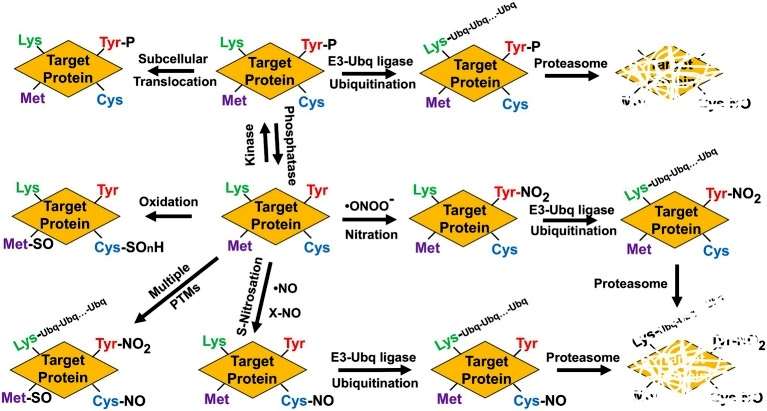 Figure 1
Figure 1
Protein nitration, a prominent post-translational modification (PTM) induced by NO, is explored in depth. The study delves into the diverse PTMs, including S-nitrosylation of cysteine residues and tyrosine nitration. The reactive nature of NO results in the formation of nitro-derivatives, including peroxynitrite, which further leads to the nitration of tyrosine, tryptophan, phenylalanine, and histidine residues. Mass spectrometry techniques are employed to identify S-nitrosylated and nitrated plant proteins, shedding light on the regulatory functions of these PTMs (Figure 2).
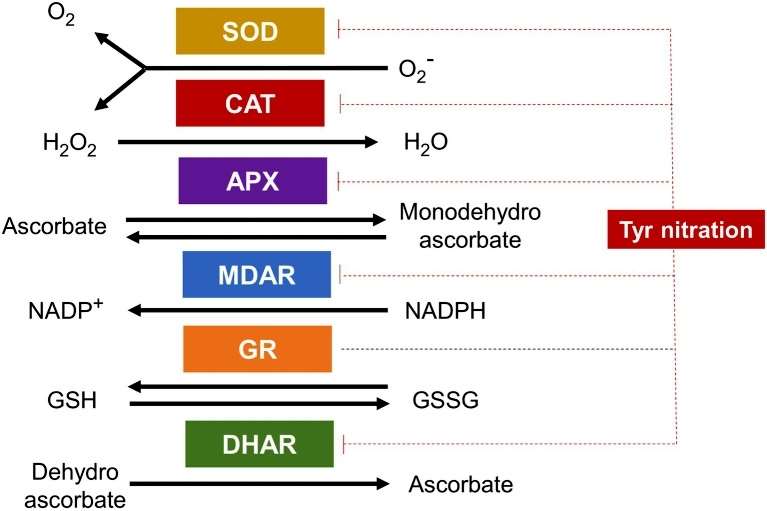 Figure 2
Figure 2
Likely the most used specific methodology to study the effects of PTMs on protein function has been the site-specific mutagenesis. Ser and Thr change to Glu or Asp has been used to mimic phosphorylation. However, this approach just allows imitating the changes in charge and steric effects caused by PTMs, but it is often far from the real PTM. A potential alternative would be the chemical synthesis of PTM-related functional groups into the protein backbone, a procedure that allows site-specific modifications of a desired protein and afford the product in large quantities for biochemical and structural analyses. This technique has been used for glycosylation, phosphorylation, ubiquitination, acetylation, and lipidation, but to our knowledge nor for nitration of S-nitrosylation. This strategy relies on the stability of the modified protein and on the chemistry used for synthesis that commonly involves the thiol group of Cys residues, which is the target residue for S-nitrosylation. An alternative method is based on the capacity of bacteria to perform genetically encoded synthesis of proteins containing non-proteinogenic amino acids including 3-nitroTyr, therefore representing a valuable platform for studying specific protein nitration effects. These genetic code expansion methodologies allow the co-translational incorporation of non-canonical amino acids into proteins (Figure 3).
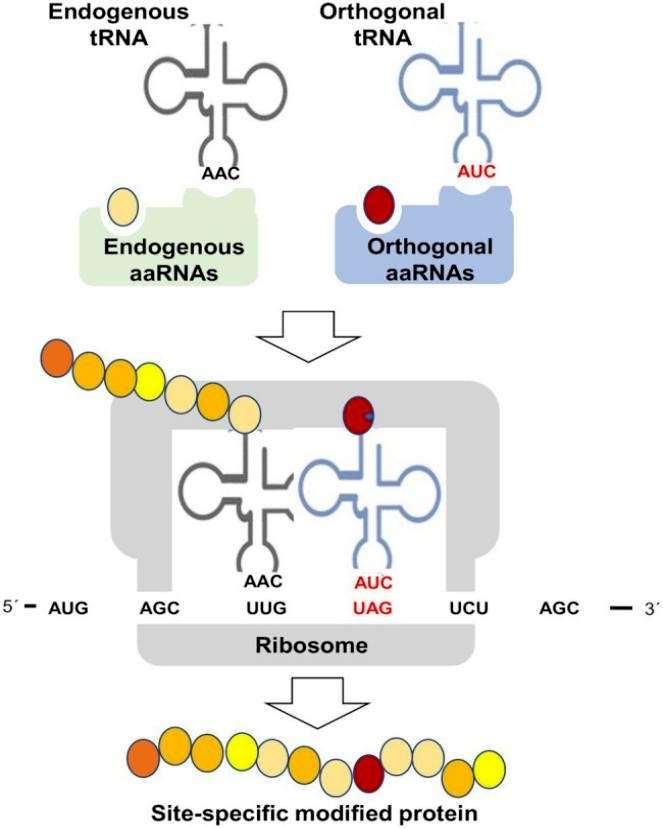 Figure 3
Figure 3
Conclusion
The study underscores the crucial role of protein nitration as a regulatory node in plant physiology, influencing processes ranging from transcriptional regulation to ROS metabolism. The results emphasize the significance of post-translational modifications, particularly tyrosine nitration, in orchestrating plant responses to environmental cues and stress conditions. The challenges associated with the identification of nitrated proteins are addressed through a multifaceted technical approach, integrating mass spectrometry, immunoprecipitation, and genetic strategies. The study calls for further exploration of genetic code expansion technologies to facilitate site-specific modification studies. Overall, the findings provide valuable insights into the complex regulatory landscape of protein nitration in plants, paving the way for a deeper understanding of its functional implications in both stress and non-stress contexts.
Reference
Our products and services are for research use only.