
- Home
- PTMs Proteomics
- Proteomics Analysis of α-Amidated Peptides
α-Amidated peptides, a class of biologically significant molecules, have garnered immense attention in the realm of biological research. These peptides, characterized by the presence of an amidated C-terminus, play pivotal roles in various physiological processes. The amidation process involves the conversion of a C-terminal glycine residue into an amide group, enhancing the stability and bioactivity of peptides.

α-Amidated peptides continue to attract researchers from a variety of disciplines with their complex structures and diverse functions. Creative Proteomics provides advanced analytical methods to help researchers advance the understanding of these peptides, revealing their role in neurobiology, endocrinology, and drug development. As emerging trends highlight their involvement in cancer biology and therapeutic modulation, the future of α-amidated peptide research holds exciting prospects for unraveling biological complexity and advancing innovative medical solutions.
A mass spectrometry-based method to screen for α-amidated peptides
Journal: Proteomics
Published: 2012
Background
α-Amidation is a crucial post-translational modification in neuropeptides, with approximately half of nervous and endocrine system peptides being α-amidated. The C-terminal amide structure is vital for the biological activity and stability of many neuropeptides. Chemical and enzymatic methods have been developed to identify α-amidated peptides, including assays based on the Hoffman rearrangement and the Tatemoto and Mutt method. Mass spectrometry (MS) and tandem mass spectrometry (MS/MS) methods have also been employed for α-amidated peptide identification, involving low energy collision-induced dissociation (CID), chemical derivatization, and immunological detection.
Results
The AtT-20 mouse pituitary corticotropic tumor cell line expresses high levels of Peptidylglycine α-amidating monooxygenase (PAM) and two amidated peptides, mouse joining peptide (mJP) and α-MSH. PAM activity in these cells is inhibited by disulfiram, leading to the accumulation of α-amidated peptide precursors. In a study using LC-MALDI-TOF-MS, it was found that the relative intensity of the precursor peptides increased significantly (2.8-fold for mJP and 9.2-fold for α-MSH) when the cells were treated with the PAM inhibitor. The reverse phase retention times of the amidated peptides and their precursors were similar, with α-amidated peptides detected in the same or neighboring fractions as the precursors. In online LC-MS/MS analysis, there were retention time differences between α-amidated and precursor peptides. The MS/MS spectra of mJP and its precursor exhibited very similar fragmentation patterns, with identical b-ion series and y ions differing by 58 Da, associated with truncation of the glycine residue and amidation of the C-terminus. These findings provide proof of concept regarding the impact of PAM inhibition on the levels of amidated peptides in AtT-20 cells (Figure 1).
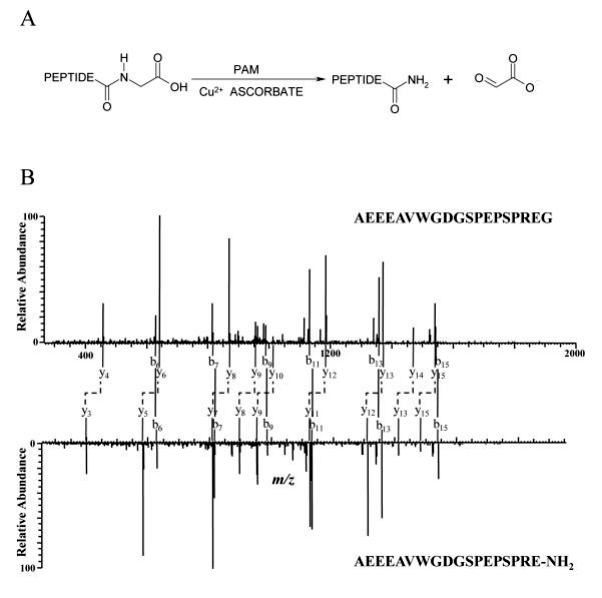 Figure 1
Figure 1
Through the identification of peaks with a mass difference of 58 between two spectra, four pairs of y ions were found with specific m/z values (458.44 and 400.30, 642.41 and 584.40, 771.46 and 713.23, 868.66 and 810.51). These y ions matched the y4, y6, y7, and y8 ions in the mJP precursor and α-amidated peptide pair, indicating that all these peptide pairs are related to mJP. The sequences, likely GSPEPSPREG and GSPEPSPRE-NH2, were inferred based on the m/z values of the parent ions. Although the complete sequences couldn't be deduced from fragments, accurate mass measurements of intact peptides compared to theoretical mass, along with assigned b ions, supported the correctness of the sequences. Similarly, another pair of peptides was identified as GDGSPEPSPREG and GDGSPEPSPRE-NH2 (Figure 2).
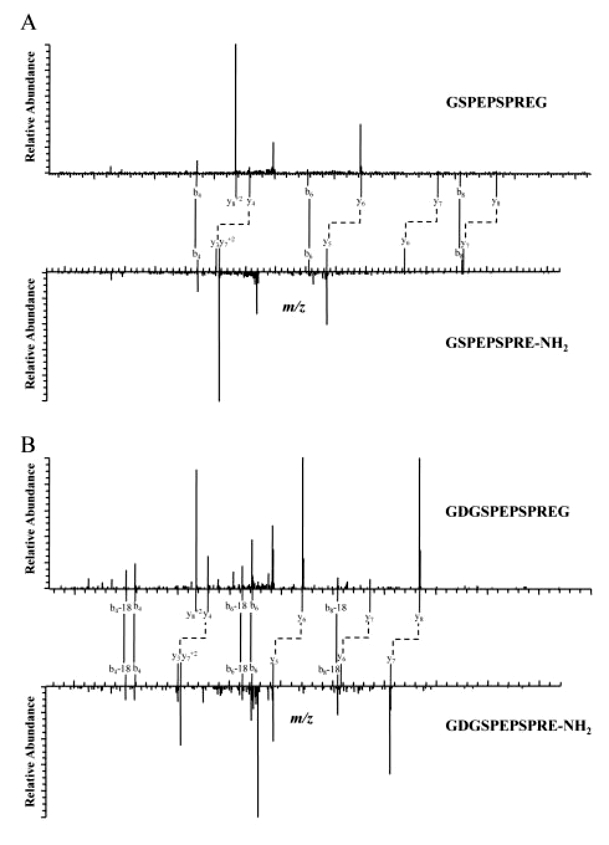 Figure 2
Figure 2
De novo sequencing was necessary to determine the sequence of the other peptide pair. Partial sequence information "EWR" or "EGER" was obtained from the peaks. The fragment ions 243.05 and 286.31 were instrumental in deducing the remaining compositions at the N-terminus and C-terminus, revealing options such as (L/I, E) or (Q/K, N) for the N-terminus and (L/I, P)G for the C-terminus. Error values in parts per million (ppm) for each possible sequence were calculated and presented in Figure 3C, taking into account the mass measurement accuracy of the mass analyzer (~5 ppm). The sequences "(Q, N) EWR(L/I, P)G" and "(L/I, E) EGER(L/I, P)G" were deemed more likely to match the actual sequence of this peptide. A BLAST search against the NCBI non-redundant protein sequences (NR) database using all possible combinations revealed one match for the sequence "ELEGERPLG." This finding was further confirmed by MS/MS of the synthetic peptide "ELEGERPL-NH2" obtained under the same conditions (Figure 3).
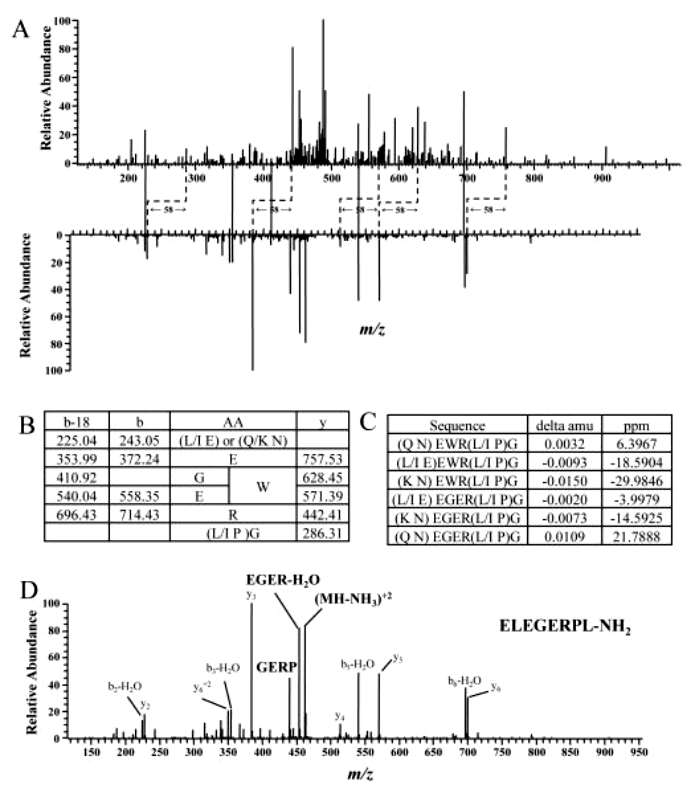 Figure 3
Figure 3
Conclusion
Peptidylglycine α-amidating monooxygenase (PAM) catalyzes this reaction, resulting in a 58 Da decrease in mass relative to the precursor peptide. The similarity in amino acid sequences of α-amidated peptides and their precursors, except for the C-terminal glycine, leads to similar reversed-phase high-performance liquid chromatography (RP-HPLC) properties and tandem mass spectral (MS/MS) fragmentation patterns. A strategy called Precursor and α-Amidated Peptide Pairing (PAPP) was developed, involving scanning LC-MS/MS data for peptide pairs differing by 58 Da but exhibiting similar RP-HPLC retention times. Growth of cultured cells in the presence of a PAM inhibitor ensured coexistence of α-amidated peptides and their precursors. The PAPP method significantly reduced the number of spectra requiring interpretation, decreasing computing time for database searching and enabling manual interpretation of unidentified spectra. The study reports the identification of α-amidated peptides from AtT-20 cells using the PAPP method.
Reference
Our products and services are for research use only.