
- Home
- PTMs Proteomics
- Post-Translational Modification of Protein Biopharmaceuticals
- Insect Cell Glycosylation Patterns Analysis
Glycosylation, the enzymatic addition of sugar moieties to proteins, profoundly influences their biological activity. In insect cells, glycosylation patterns contribute significantly to the functionality of expressed proteins, impacting aspects such as receptor binding, immune response modulation, and therapeutic efficacy. Insect cells exhibit both N-linked and O-linked glycosylation pathways. N-glycosylation involves the attachment of glycans to asparagine residues, while O-glycosylation decorates serine or threonine residues with sugar moieties. Understanding the interplay between these pathways is crucial for deciphering the glycosylation landscape in insect cells.
Insect cells have emerged as valuable hosts for recombinant protein expression due to their efficient protein folding machinery and post-translational modification (PTM) capabilities. Among these PTMs, glycosylation plays a pivotal role in determining the structure, function, and stability of proteins.
Mass Spectrometry (MS) Approaches
Glycan Profiling Techniques

The analysis of insect cell glycosylation patterns is a multifaceted endeavor with far-reaching implications in biotechnology and medicine. Creative Proteomics stays abreast of advanced analytical technologies and continues to provide effective solutions for unlocking the ability to unravel the complex language of insect cellular glycosylation and facilitating innovation in the design and production of therapeutic proteins.
Site-specific O-glycosylation analysis of SARS-CoV-2 spike protein produced in insect and human cells
Journal: bioRxiv
Published: 2021
Background
Enveloped viruses hijack not only the host translation processes, but also its glycosylation machinery, and to a variable extent cover viral surface proteins with tolerogenic host-like structures. SARS-CoV-2 surface protein S presents as a trimer on the viral surface and is covered by a dense shield of N-linked glycans, and a few O-glycosites have been reported. The location of O-glycans is controlled by a large family of initiating enzymes with variable expression in cells and tissues and hence difficult to predict. Here, we used our well-established O-glycoproteomic workflows to map the precise positions of O-linked glycosylation sites on three different entities of protein S - insect cell or human cell-produced ectodomains, or insect cell derived receptor binding domain (RBD).
Results
The study involved the expression of pre-fusion stabilized S ectodomains in Drosophila S2 and human embryonic kidney (HEK) 293F cells, utilizing two slightly different constructs. Both constructs included a mutated S1/S2 cleavage site and stabilizing mutations (986P, 987P) in the S2 region. The human cell product, featuring a C-terminal leucine zipper (GCN4) motif, primarily existed as a trimer, while the insect cell product lacked a forced trimerization motif, existing as several entities, predominantly as a dimer based on size exclusion chromatography. Additionally, a soluble monomeric receptor binding domain (RBD) was expressed in insect cells.
Mucin type O-glycosylation sites on the recombinantly expressed S proteins were mapped using an established ETciD and HCD nLC-MS/MS workflow on a Thermo Fisher Orbitrap Fusion Lumos Tribrid mass spectrometer. An in-gel digestion approach with chymotrypsin or Glu-C followed by trypsin digestion was employed, with PNGase F treatment to remove N-glycans. The study aimed to identify unmodified and de-N-glycosylated peptides through deamidation of asparagine. The combined digestion strategy achieved high sequence coverage, with over 96% of individual sequences identified. Chymotrypsin digestion yielded higher numbers of peptide groups and glycoPSMs, indicating improved separation or sensitivity. Neuraminidase treatment focused on identifying simple GalNAcα-S/T (Tn) and Galβ3GalNAcα-S/T (core 1) structures, prevalent in both HEK 293F and S2 cell lines, with core 1 structures usually capped with sialic acid in human cells and unmodified in insects (Figure 1).
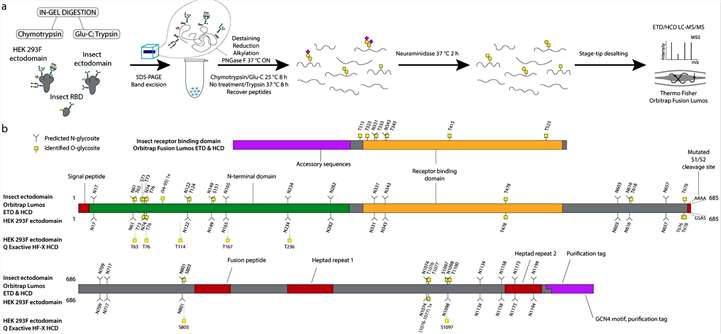 Figure 1
Figure 1
The author analyzed data derived from the Orbitrap Fusion Lumos and Q Exactive HF-X mass spectrometers to assess O-glycosylation on recombinantly expressed S proteins. The findings indicated low occupancies at most sites, with less than 5% of quantified peptides carrying O-glycans. The highest occupied O-glycosites were identified near specific N-glycosylation sites. Notably, many O-glycosites were located within or just before N-X-S/T sequons of N-glycosylation. To investigate coexistence with N-glycans, N-X-S/T covering O-glycopeptides were split into two groups: those with asparagine (non-N-glycosylated) and those with aspartic acid (suggestive of deamidation after PNGase F treatment). Most O-glycosites near N-X-S/T sequons were found on non-N-glycosylated peptides, with few on N-deamidated peptides and minimal occurrences on peptides without O-glycans. In 7 out of 9 glycan clusters of interest, over 85% of quantification signals came from O-glycopeptides when no N-glycans were present. For Q Exactive HF-X data, peptides of different lengths covering O-glycosites were grouped, and occupancy was calculated. Similar to the Orbitrap data, very low occupancies were estimated for commonly identified O-glycosites (Figure 2).
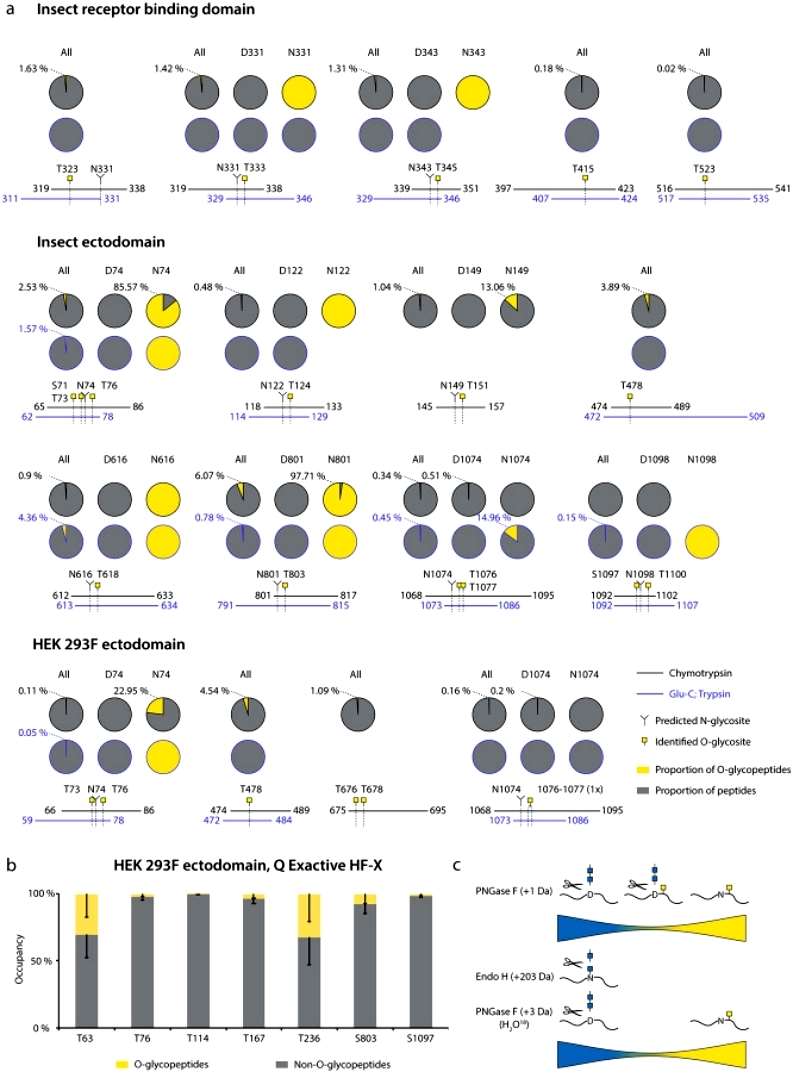 Figure 2
Figure 2
To visualize the 3D context of O-glycosites, Galβ3GalNAcα disaccharides were manually attached to the 3D model and optimized using YASARA. The resulting structures were trimmed to GalNAcα for visualization, especially if only such structures were identified by mass spectrometry. This approach provided insights into the spatial arrangement of O-glycosylation on the S protein structure (Figure 3).
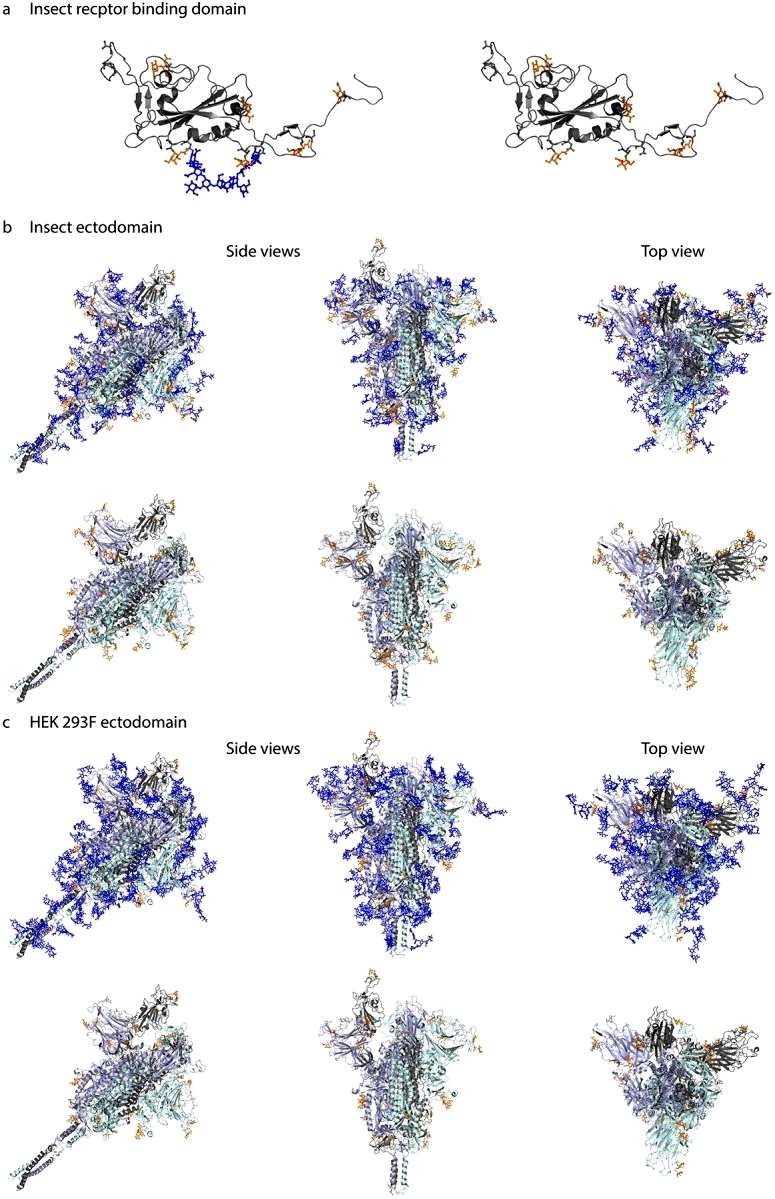 Figure 3
Figure 3
Conclusion
The article utilized established O-glycoproteomic methods to precisely locate O-linked glycosylation sites on three distinct protein S entities: ectodomains produced in insect cells or human cells, and the receptor binding domain (RBD) derived from insect cells. The study identified 25 O-glycosites, revealing similar patterns in the ectodomains from different cell origins but a distinct pattern in the monomeric RBD. Notably, 16 out of 25 O-glycosites were situated within three amino acids of known N-glycosites. However, O-glycosylation was predominantly observed on peptides without N-glycans, suggesting potential complementary roles of O-glycans in immune shielding. The study implies minimal impact of O-glycosylation on subunit vaccine design for SARS-CoV-2.
Reference
Our products and services are for research use only.