
- Home
- PTMs Proteomics
- Protein PTMs Site Identification
Post-translational modifications (PTMs) encompass chemical alterations occurring subsequent to protein translation. Extensive research has unveiled a diverse array of PTM types, with prevalent investigations focusing on phosphorylation, acetylation, methylation, glycosylation, ubiquitination, among others. These PTMs substantially enhance the intricacy of the proteome, exerting profound effects on protein conformation, distribution, and functionality. Their significance in cell biology is paramount, spanning vital roles in cell signaling, structural organization, DNA modifications, and more. Notably, PTMs of a given protein in distinct physiological states, subcellular compartments, and amino acid loci can confer varying functions. Consequently, protein post-translational modification remains an active focal point and challenge in contemporary scientific inquiry.
In this context, mass spectrometry emerges as a potent tool for scrutinizing protein modification sites. This technology enables the precise identification of modification sites within individual proteins and facilitates high-throughput analysis of proteome-wide modification profiles.
Within the domain of protein post-translational modifications, particular residues in a protein's amino acid sequence are susceptible to covalent interactions with chemical entities or low-molecular-weight proteins. Such covalent conjugation events lead to readily observable modifications in the molecular weight of the implicated protein sequence. In the contemporary scientific landscape, the detection and characterization of specific protein PTMs have been significantly facilitated by the utilization of advanced mass spectrometry techniques. The application of high-precision mass spectrometry enables the precise differentiation of molecular weight variations between unmodified and modified protein states.
This capability allows us to undertake a meticulous dissection of the molecular weight shifts that transpire before and after post-translational modification events affecting a target protein. Through the amalgamation of cutting-edge mass spectrometry methodologies and artificial intelligence-driven data analysis, we can provide a comprehensive and precise framework for the identification and quantification of a wide spectrum of PTMs, thereby advancing our understanding of the intricate landscape of protein regulation and function.
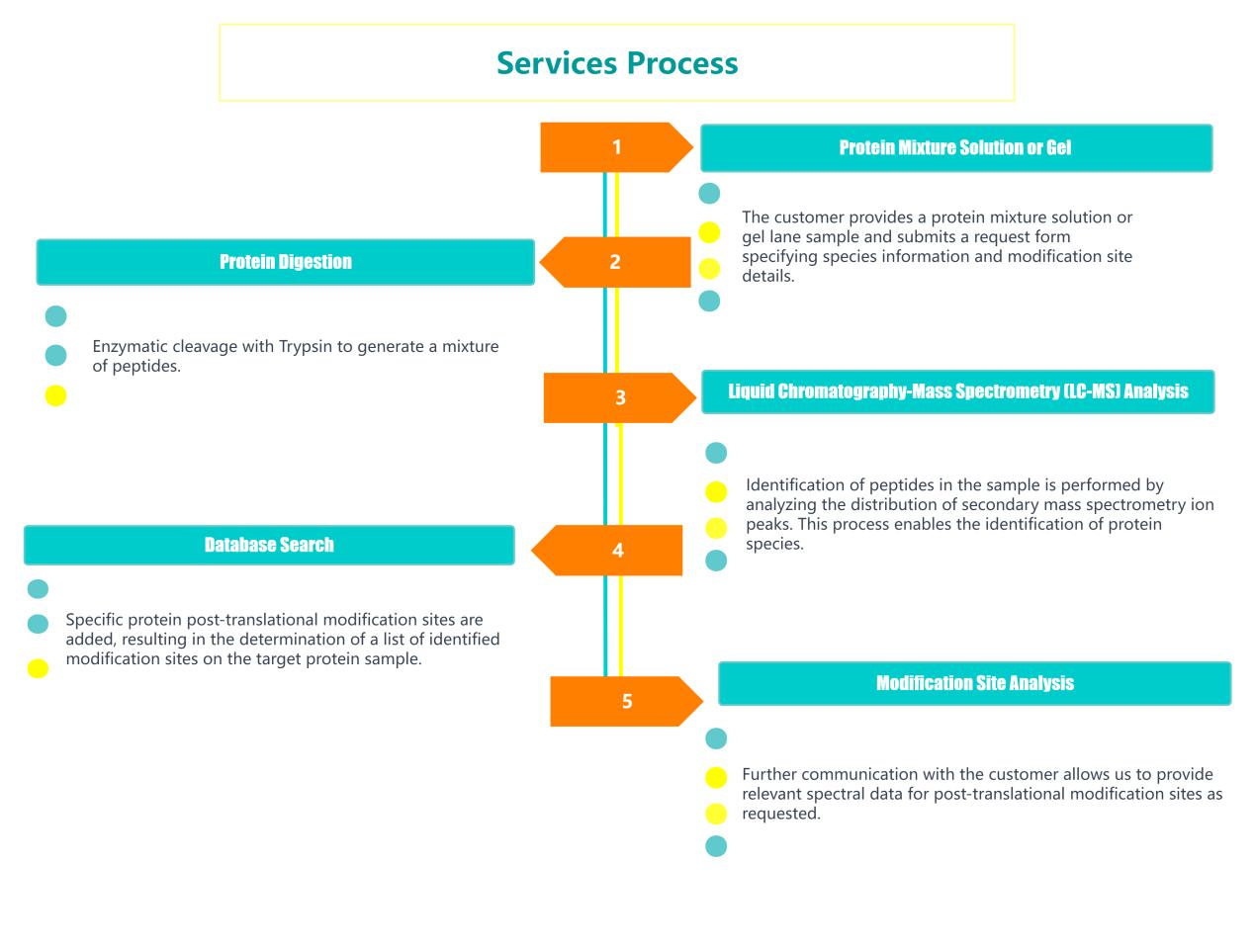
Identification of protein post-modification sites refers to the peptides that have been modified after modification in the IP samples, protein mixture samples, and SDS-PAGE lane strip samples provided by the customer through liquid mass spectrometry (LC-MS/MS) technology. Mass spectrometry detection, combined with relevant species database searches, can detect various currently known protein modification types and some small molecule drugs to analyze the modification sites on the protein to find out the exact modification site of the peptide, and Provide mass spectrum information of specific modification sites.
Using SDS-PAGE and liquid chromatography-tandem mass spectrometry (LC-MS/MS), Creative Proteomics locates modification sites on single proteins acquired through immunoprecipitation (IP). This makes it possible to identify all recognized modification types on proteins as well as the precise modification sites of particular small molecule medications.
phosphorylation, glycosylation, methylation, acetylation, ubiquitination, propionylation, butyrylation, malonylation, Glutarylation, succinylation.)
There are many types of protein modifications that can be detected. Protein ubiquitination modifications, phosphorylation modifications, and glycosylation modifications can be identified through database searches.
Pre-processing does not require post-modification enrichment. Conventional post-modification site detection does not require enrichment processing of samples and can identify multiple post-modifications at the same time.
High instrument stability and short cycle time: advanced mass spectrometry instruments, mature operators, high identification accuracy and sensitivity.
Three general modification types can be scanned at once.
The analysis is not limited to conventional PTM, and personalized targeted modification analysis can be performed based on the determined amino acid positions and modification types.
| Data Analysis Content | Description |
|---|---|
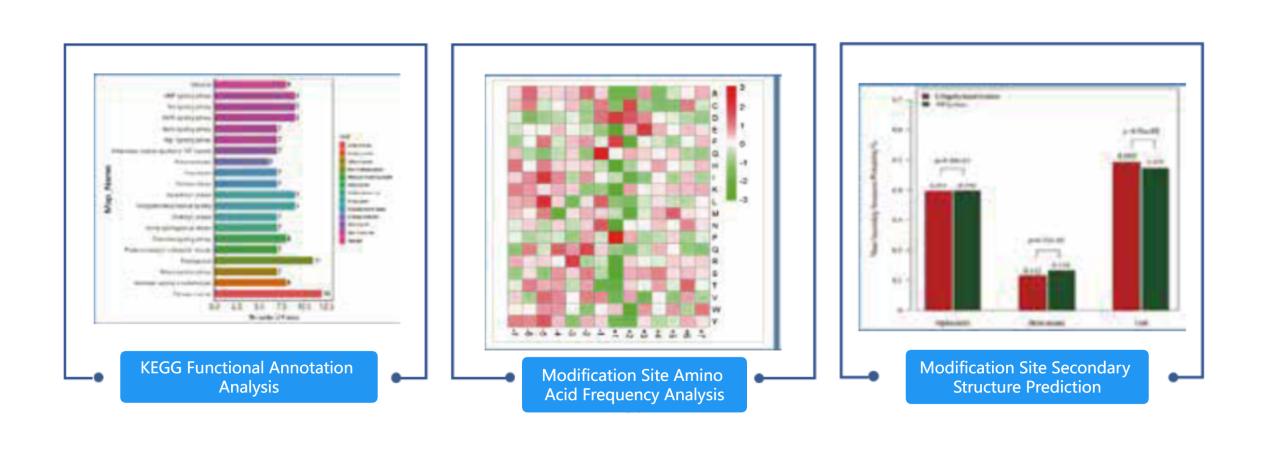
| Modification Expression Profile Analysis | Identification Results Statistics: Protein identification result statistics, modification site amino acid frequency distribution, modification site secondary structure prediction, motif analysis. |
| Biological Function Analysis: Subcellular localization, domain analysis, GO functional analysis, KEGG pathway analysis, interaction network, and module analysis. | |
| Identification of Modified Proteins | Biological Function Analysis: GO annotation analysis (limited to Uniprot database only). |
SDS-PAGE Gel Lane: Visible silver staining or Coomassie staining bands.
Protein Solution: Total protein amount > 5μg, concentration > 0.1μg/μl.
Thermo Scientific Q Exactive
Identification of single protein modifications.
Identification of modification sites for small molecule drug-protein interactions.
Protein modification identification in low-complexity samples.
Our products and services are for research use only.