
- Home
- PTMs Proteomics
- Global PTMs Profiling Service
Post-translational modifications (PTMs) of proteins encompass a regulatory mechanism wherein precise chemical groups are introduced to or removed from amino acid residues. This intricate process intricately governs protein functionality, subcellular localization, expression dynamics, and interplay with other cellular constituents.
To date, more than six hundred types of protein modifications have been described, including well-known ones such as phosphorylation, ubiquitination, glycosylation, methylation, acetylation, SUMOylation, short-chain and long-chain acylation, oxidative-reduction modifications, ADP-ribosylation, neddylation, glutaminylation, and others. Most PTMs are dynamic and reversible, occurring more rapidly than the synthesis of new proteins, enabling cells or organisms to respond rapidly to changes in their environment. Increasingly, research has revealed that many essential biological processes and disease occurrences are not only correlated with protein abundance but, more importantly, regulated by various protein PTMs.
Global PTMs Profiling is of significant importance in uncovering the mechanisms of life processes, identifying clinical biomarkers for diseases, and pinpointing drug targets. By delving into the differential changes at the level of protein post-translational modifications, proteomics sheds light on various aspects of life activities.
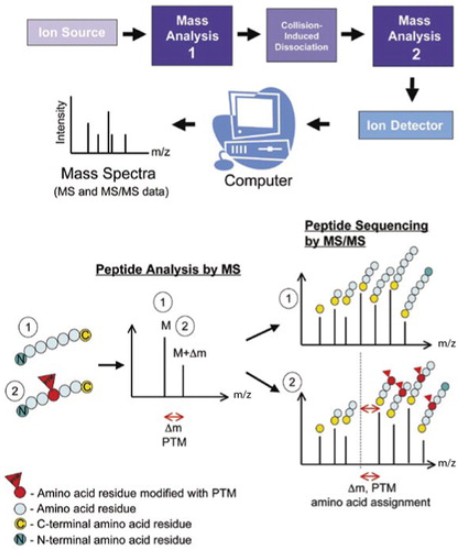 Fig. 1 Tandem mass spectrometry (MS/MS) for mapping posttranslational modifications (PTMs). (Larsen, Martin R., et al., 2006)
Fig. 1 Tandem mass spectrometry (MS/MS) for mapping posttranslational modifications (PTMs). (Larsen, Martin R., et al., 2006)
Liquid Chromatography-Mass Spectrometry (LC-MS/MS) technology is the preferred method for identifying protein modifications. It allows simultaneous identification of modification types, modification sites, and relative quantification of modifications. This article primarily introduces the principles of mass spectrometry-based identification of post-translational protein modifications. It also analyzes the differences between modification identification and modification proteomics products, as well as the associated technical challenges.
Based on effective protein/peptide enrichment strategies, advanced MS instrumentation, and proteomics technologies, Creative Proteomics can help our global customers to perform fast and reliable deep protein PTM profiling.
Our Profiling PTMs Solution globally involves identifying and quantifying a wide range of PTMs simultaneously. This service enables the identification, localization and quantitation of the proteins that are post-translationally modified as well as the type of post-translational modification.
| Method | Application |
|---|---|
| Modification Identification Analysis | Identification of modification sites |
| Qualitative Analysis | Proteomic analysis of protein modifications |
| Quantitative Analysis | Quantitative profiling of modified proteomes |
| Absolute Quantification of Modifications | Absolute quantification of modified proteomes (PRM) |
The deliverables include a comprehensive written report that provides detailed information about the samples, our methods, and a summarized representation of the data.
Additionally, an Excel workbook will be provided, which entails both qualitative and quantitative protein identification data in a meticulous manner.
Bioinformatics analysis includes: 1. Identification of modified proteins and modification sites; 2. Characteristic distribution of identification results: peptide length, protein coverage distribution, unique peptide distribution; 3. Modification site motif prediction; 4. Protein GO functional classification, COG annotation, Pathway annotation; 5. Protein network interaction prediction
We have developed a distinctive bioinformatics analysis workflow aimed at extracting biologically relevant information from complex experimental data. Through integrated analyses of proteomic data, we provide our clients with insights into protein expression levels and functional regulation beyond protein expression.
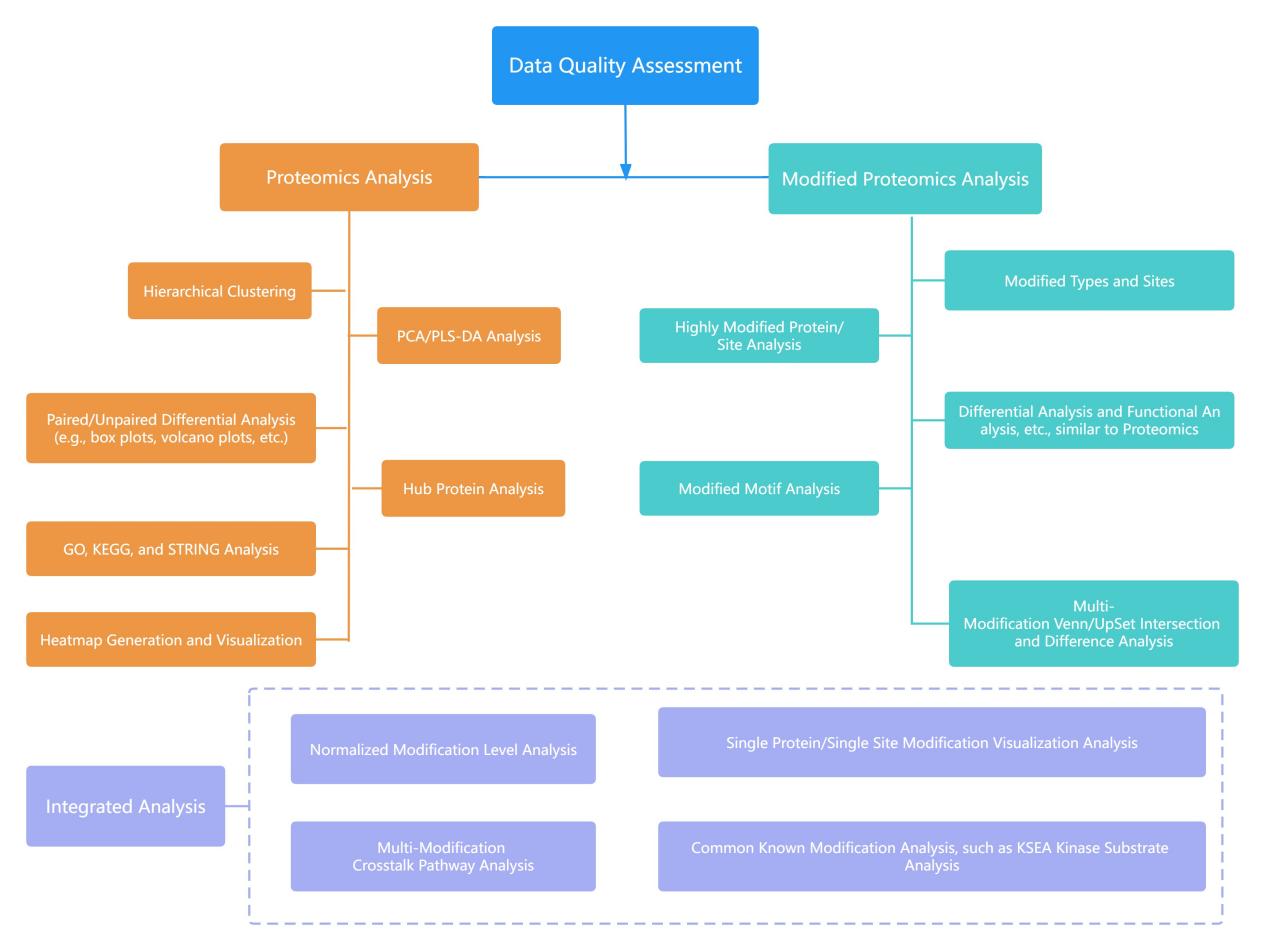
The content of the modification proteomics data analysis includes:
Statistical analysis of search results and data quality assessment.
Proteomic analysis such as hierarchical clustering analysis, PCA/PLS-DA analysis, differential analysis based on sample types, and common pathway annotation analysis, including GO analysis, KEGG analysis, protein-protein interaction analysis, and identification of hub proteins.
Statistical analysis of the pan-modification group and descriptive statistics of highly modified protein sites. Additional differential and functional analysis is carried out on the whole proteome. This also includes Motif analysis of modification features and the intersection and difference analysis of modification pathways and proteins with multiple modifications.
Analysis of protein co-modification groups, including pathway analysis of cross-talk in multi-modifications, visualization of single protein/site modifications, and common modification analyses such as KSEA analysis.
Data Process
In-Depth Analysis of Diverse Modification Types: Post-translational proteomics facilitates a comprehensive exploration of a wide array of protein post-translational modifications (PTMs), encompassing phosphorylation, acetylation, ubiquitination, methylation, and other modifications. This approach unveils the multifaceted functionalities of proteins. Notably, the analysis is not restricted to conventional PTMs and can be fine-tuned for targeted modification analysis based on specific amino acid residues and modification categories.
High-Throughput Capabilities: This methodology supports high-throughput processing of samples, allowing for the simultaneous analysis of multiple samples. This expedites the pace of research and enhances experimental efficiency.
Exceptional Sensitivity and Precision: Employing high-resolution mass spectrometry for analysis, this technology offers remarkable sensitivity and precision. It can discern modified proteins with low abundance and provide dependable quantitative data.
Robust Large-Scale Data Handling: Post-translational proteomics demonstrates robust capabilities in handling extensive data sets, accommodating intricate sample profiles, and identifying numerous modification sites. This capacity furnishes comprehensive information, thereby facilitating in-depth exploration of protein functions and regulatory mechanisms.
Customized Experimental Designs: Customized experimental procedures can be created to meet specific research goals. This adaptability enables the examination of various sample types and modification categories, effectively satisfying the particular needs of different research endeavors.
Sample Preparation: The initial phase encompasses the extraction of proteins from the specific biological sample of interest, which could encompass cells, tissues, or biofluids. Subsequently, the sample undergoes proteolysis, resulting in the breakdown of proteins into peptide fragments. Distinct proteolytic enzymes such as trypsin or Lys-C may be utilized for this purpose.
Enrichment of Modified Peptides: Since post-translational modifications (PTMs) are frequently present at low stoichiometry within the proteome, specialized enrichment techniques are employed to selectively isolate peptides harboring specific modifications. For instance, in the investigation of phosphorylation, methodologies such as immobilized metal affinity chromatography (IMAC) or titanium dioxide (TiO2) chromatography are employed. Various other enrichment strategies are also available for diverse PTMs.
Mass Spectrometry (MS): The enriched peptide fractions are subjected to analysis using mass spectrometry, a potent analytical technique for determining the mass-to-charge ratio of ions. High-resolution MS instruments, including Orbitrap or Triple Quadrupole systems, are commonly employed for the profiling of PTMs.
Data Acquisition: During the mass spectrometry analysis, the instrument records mass spectra of the peptides. These spectra furnish critical information regarding the peptide's mass, charge, and fragmentation pattern, facilitating the subsequent identification and quantification of PTMs.
Database Searching and Identification: Proficient bioinformaticians analyze the mass spectrometry data utilizing comprehensive databases encompassing known protein sequences and PTMs. This analysis aids in the identification of the peptides and the discernment of their associated modifications.
Quantitative Analysis: Quantitative insights pertaining to the abundance of modified peptides are furnished. This may encompass approaches such as label-free quantification or the utilization of isobaric labeling techniques such as Tandem Mass Tags (TMT) or Isobaric Tags for Relative and Absolute Quantification (iTRAQ).
Bioinformatics Analysis: A comprehensive suite of bioinformatics tools is harnessed to extract meaningful biological information from the PTMs data. This encompassing analysis includes:
Functional enrichment analysis, facilitating the identification of enriched pathways and biological functions.
Network analysis, providing insights into the interactions among modified proteins.
The deployment of visualization tools to enable researchers to interpret the data effectively.
Data Reporting: The culminating phase involves the generation of a detailed report that encapsulates the identified PTMs, their respective abundance levels, statistical analyses, and biological implications. This report is often complemented by graphical representations and supplementary information.
Creative Proteomics strives to be a guide and assistant in protein PTM research for our customers worldwide. Based on the needs of our customers, we provide the best analytical strategies and solutions to facilitate biomarker discovery, disease research, protein biopharmaceutical characterization, and more. For more information on our PTM proteomics analysis services, please use our contact form. Our customer service representatives are available 24 hours a day, from Monday to Sunday.
Reference
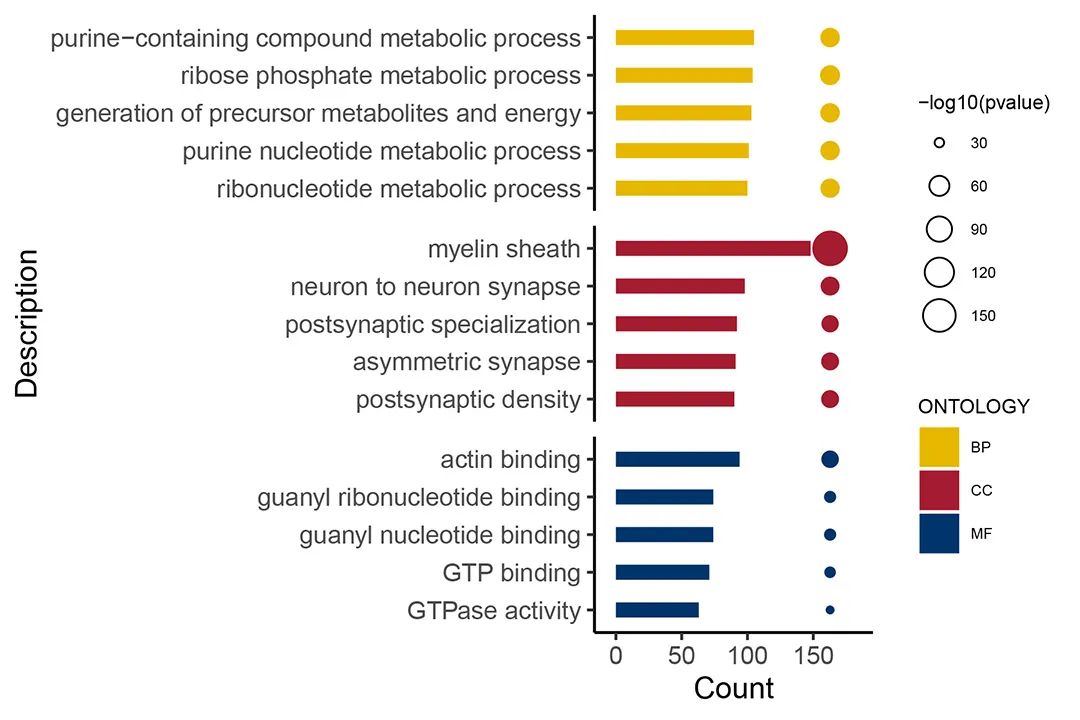
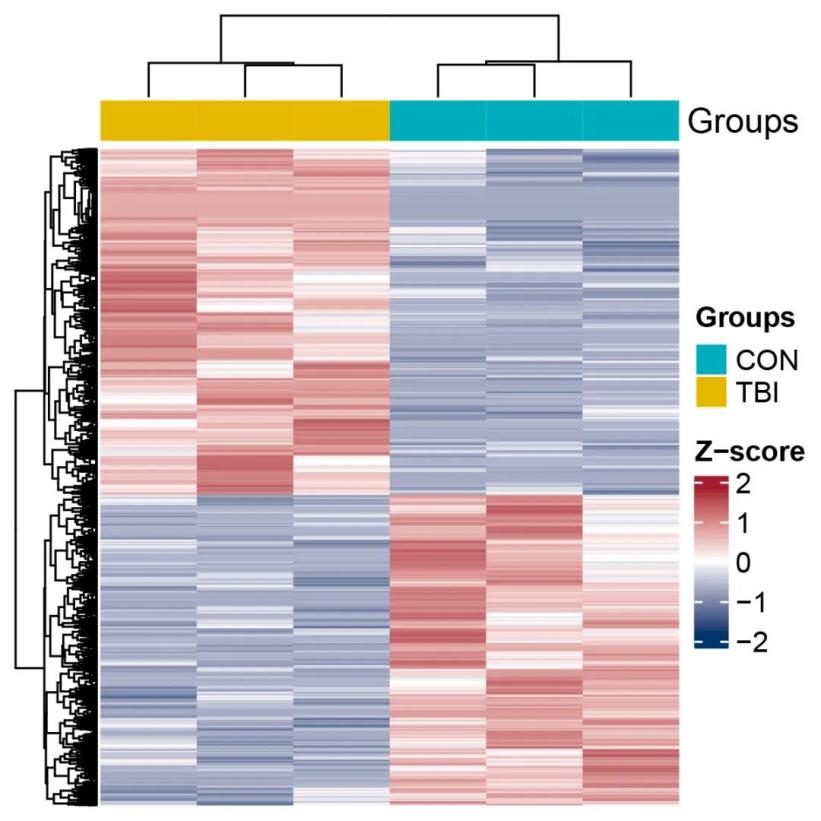
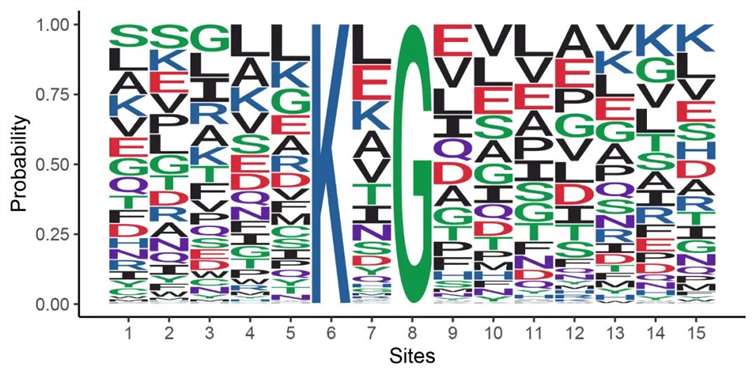
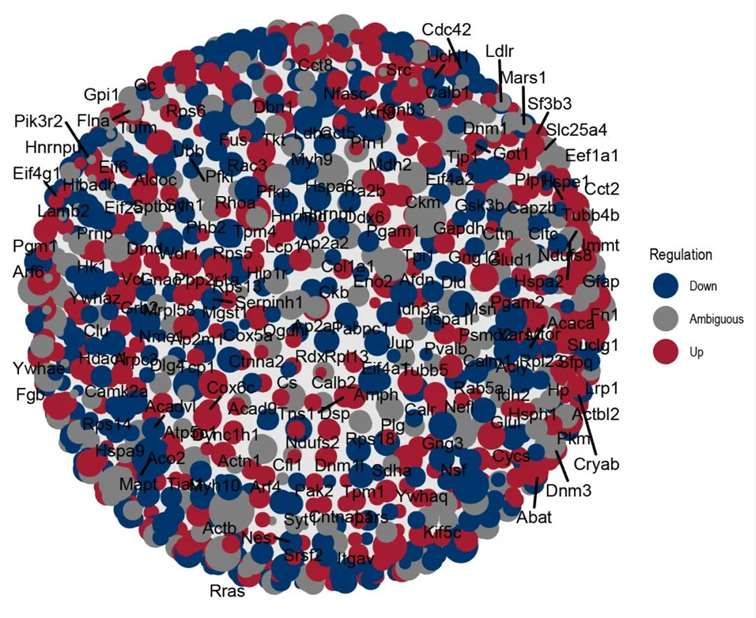
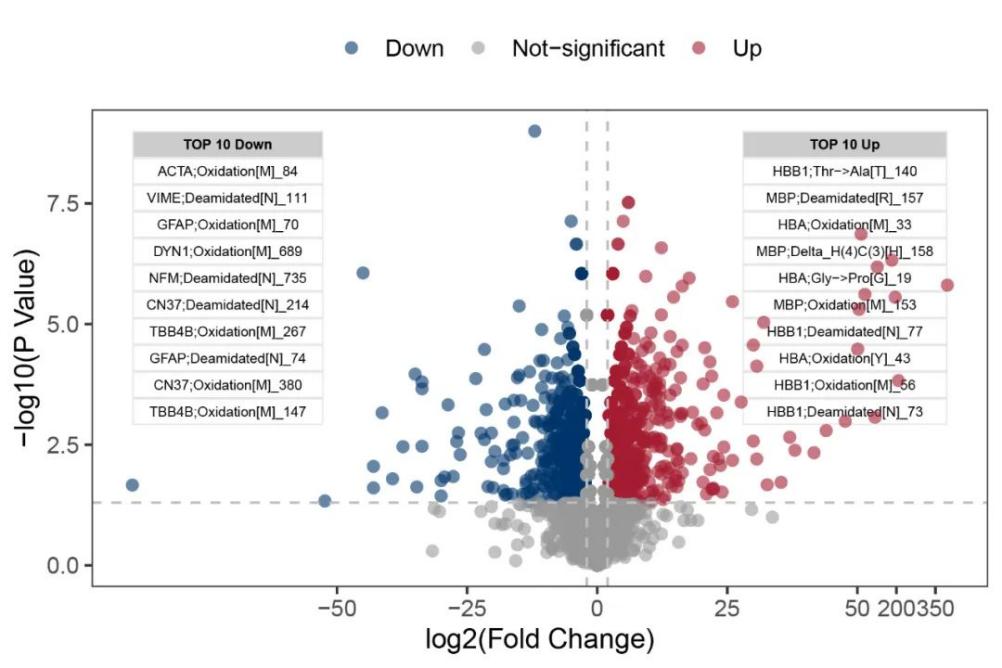
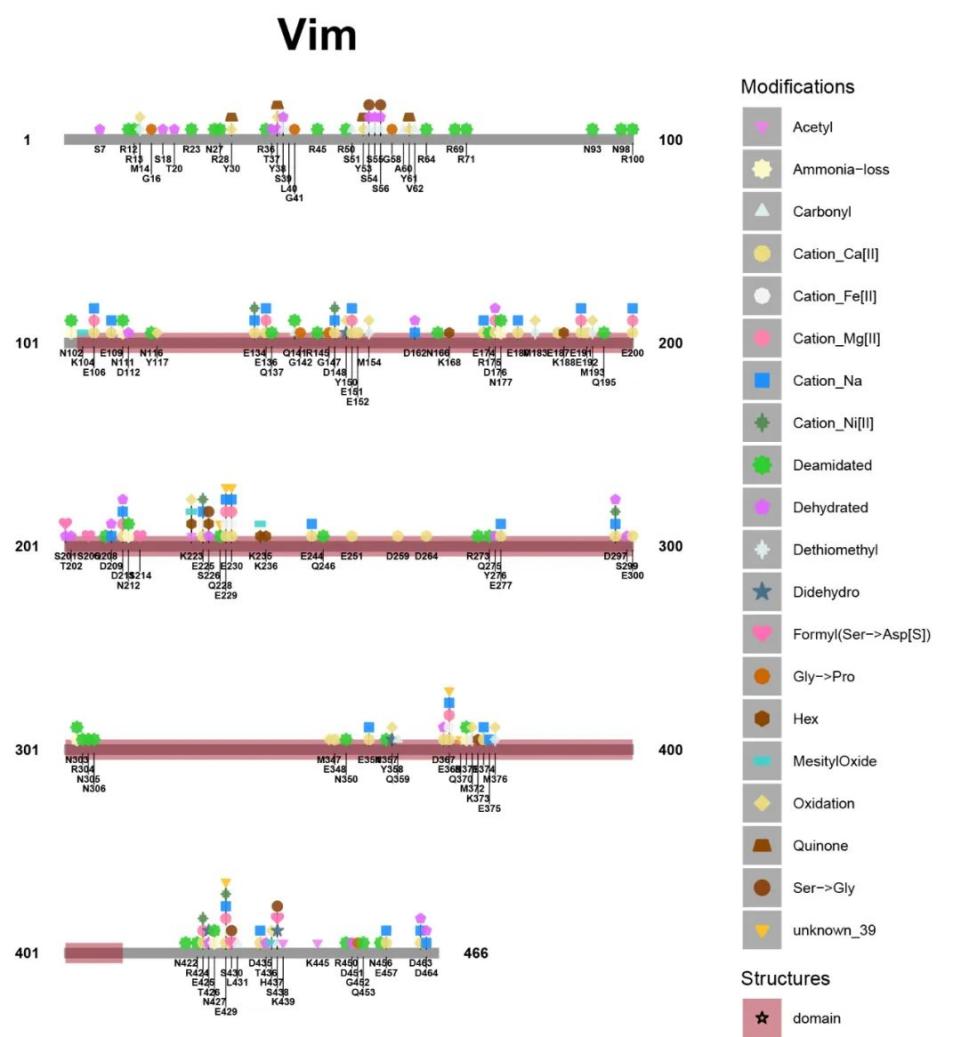
Partial Analysis Graph Examples
Our products and services are for research use only.