
- Home
- PTMs Proteomics
- PTM Quantitative Analysis
- SILAC-Based Quantitative PTM Analysis
Mass spectrometry (MS)-based analysis is a powerful technique in the unbiased study of post-translational modifications (PTMs). In addition to the qualitative annotations of PTMs, the MS-based methods can also provide quantitative information on PTMs, including their relative abundance and dynamic changes. To help our global customers achieve accurate PTM profiling across multiple biological samples, Creative Proteomics is proud to offer stable isotope labeling by amino acid in cell culture (SILAC)-based quantitative PTM analysis service, one of the most popular labeling techniques for MS-based quantitative proteomics. The application of SILAC is mainly limited to actively dividing cell lines, allowing precise quantification of PTM changes between different functional states.
As an in vitro labeling technique, SILAC is primarily used to label peptides by culturing organisms in cultures containing natural (light) or isotope-encoded (heavy) amino acids. During the liquid-chromatography-coupled high-resolution tandem MS (LC-MS/MS) analysis, isotope mass shifts between different isotopically labeled samples make it easy to distinguish between source samples. Therefore, the variation introduced from sample processing is minimized. SILAC has several advantages over the label-free and chemically labeled quantification strategies used in proteomics research, including straightforward and relatively easy implementation, high quantitative accuracy, and good reproducibility. Thus, this metabolic labeling strategy allows superior quantitative analysis of the cellular proteome and specific PTMs.
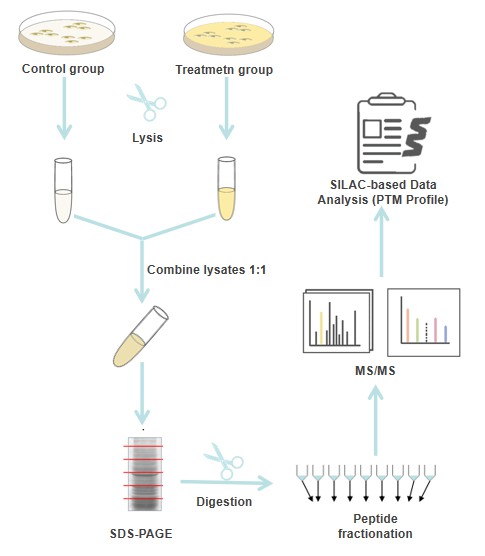
SILAC is widely used for the characterization of proteomic changes among different biological samples, the study of dynamic changes in PTMs, the differentiation of specific interacting proteins in interaction proteomics analysis, and protein turnover analysis at the proteomic scale. At Creative Proteomics, we offer a one-solution SILAC-based quantitative PTM analysis service, including cell treatment, lysis and SDS-PAGE, MS analysis, and quantification of site-specific stoichiometry of PTMs. In addition to a high-throughput SILIA-based quantitative PTM analysis to profile changes in protein PTMs, we have SILAC data analysis and PTM profile capabilities to support complex SILAC data analysis. Based on our experienced team of experts and statistical tools, we can greatly assist researchers and professionals in facilitating SILAC data analysis.
Creative Proteomics is dedicated to providing the optimal analysis strategies and solutions according to the specific project requirements of our clients. Feel free to contact us for more information about our services. We are always available for your inquiries!
References
Our products and services are for research use only.