
- Home
- PTMs Proteomics
- Proteomics Analysis of Succinylation
Lysine succinylation modification is a novel post-translational modification (PTM) of histones that are commonly found in the cytoplasm and nucleus of all prokaryotes and eukaryotes, participates in and regulates biological processes in almost all organisms, and is closely associated with diseases. Creative Proteomics specializes in the analysis and identification of PTMs based on high-resolution mass spectrometry (MS), aiming to provide quality and efficient solutions for our customers' research projects. Here, we offer protein succinylation analysis services, including protein extraction and proteolysis, peptide enrichment and separation, liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, MS raw data analysis, and bioinformatics analysis.
Lysine succinylation, defined as the transfer of a succinyl group to a lysine residue of a protein, is dynamic, reversible, and evolutionarily conserved and has been extensively studied in both mammalian and bacterial cells. MS-based proteomic analysis identified a large number of lysine succinylated-cytosolic, mitochondrial and nuclear proteins, taking part in various cellular and biological processes, such as fatty acid oxidation, metabolic enzymes for fatty acid synthesis, amino acid degradation, and mitochondrial respiration. In addition, emerging clinical studies have reported that dysregulation of protein succinylation is associated with cardiovascular diseases, cancer, and other diseases. Therefore, an in-depth understanding of succinylation modification and its regulatory mechanisms is important for understanding its physiological functions as well as for developing drug therapies.
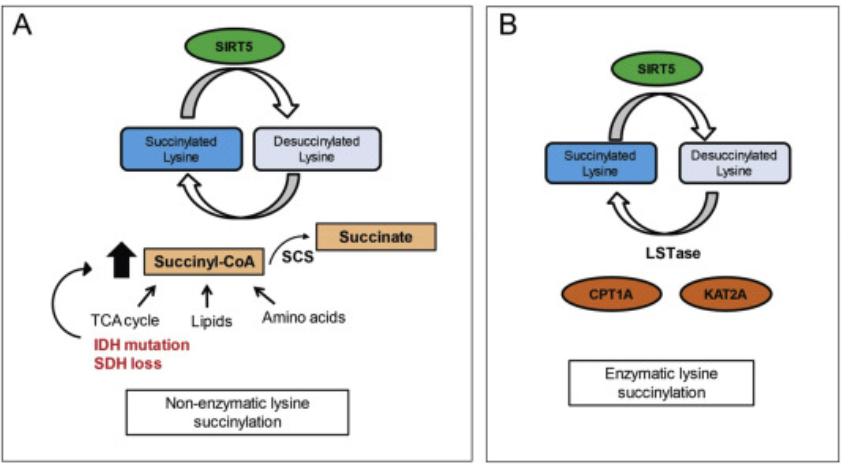 Fig. 1
Metabolic regulation of Ksucc by succinyl-CoA (A). Enzymatic regulation of Ksucc (B). (Sreedhar, et al.,
2020)
Fig. 1
Metabolic regulation of Ksucc by succinyl-CoA (A). Enzymatic regulation of Ksucc (B). (Sreedhar, et al.,
2020)
With advanced experimental techniques, Creative Proteomics is dedicated to offering fast and accurately succinylated proteins and lysine succinylation sites. We are able to characterize lysine succinylation, the distribution of modification sites, and the main influencing factors. Specifically, our service includes the following lists:
-Protein annotation.
-KEGG pathway annotation.
-GO/KEGG pathway functional enrichment analysis.
-Analysis of sequence model around succinylation sites.
-Motif-based clustering analysis.

As a newly discovered PTM, succinylation not only triggers more changes in protein properties but also has more significant effects on protein structure and function because of the larger spatial structure of the succinyl group. If you are interested in our protein succinylation analysis, please feel free to contact us. We are ready to assist you!
References
Our products and services are for research use only.