
- Home
- DNA/RNA Modification
- tRNA Modification LC-MS Analysis Service
tRNA (transfer RNA) is a small non-coding RNA that consists of both nuclear and mitochondrial tRNAs. It has a length of approximately 75-90 nucleotides and exhibits high conservation across species. tRNA plays a crucial role in mRNA decoding and protein translation, serving as a core component of these processes.
Mature tRNA possesses a well-defined secondary structure, which includes the D-loop, T-loop, anticodon loop, and CCA amino acid acceptor arm. The 3'-OH end of tRNA carries a specific amino acid, and through codon-anticodon pairing, it translates the nucleotide sequence information of mRNA into the amino acid sequence information of proteins.
To date, tRNA is the most extensively modified RNA molecule discovered, with a wide variety and abundance of post-transcriptional modifications. These modifications are crucial for various core aspects of tRNA function, such as folding, stability, and decoding. Generally, modifications on the body of tRNA are essential for tRNA structure folding, stability, rigidity, and flexibility (e.g., m7G/m5C), while modifications in the anticodon loop influence protein translation through effects on open-loop structure, codon-anticodon pairing, wobble effects, and prevention of translational frameshifting (e.g., mcm5s2U/mcm5U). Furthermore, tRNA modifications can affect aminoacylation mediated by aminoacyl-tRNA synthetases (aaRS), thereby impacting the accuracy of amino acid recognition. In general, tRNAs with low modifications are prone to degradation, making the investigation of the relationship between tRNA modification changes and tRNA expression changes an important research direction.
Defects and alterations in tRNA modifications and related modifying enzymes are associated with various human diseases, including cancer, diabetes, neurodegenerative syndromes, cardiovascular diseases, and mitochondrial-related disorders. Based on this, the analysis of tRNA modification profiles is crucial for establishing links between diseases, tRNA modifying enzymes, and tRNA molecular functions.
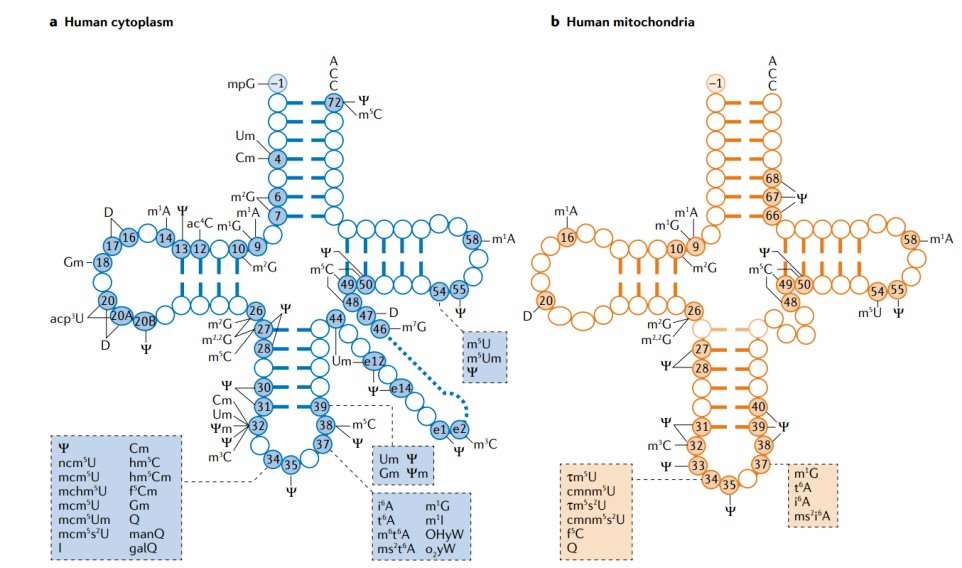 Figure 1: Summary of common types of tRNA modifications.
Figure 1: Summary of common types of tRNA modifications.
To detect the changes in the abundance of different modifications on tRNA, Creative Proteomics offers a one-stop solution for LC-MS analysis of tRNA modification quantification. This service provides analysis of 56 types of tRNA nucleoside modifications, starting from total RNA as the input material. The workflow involves the extraction of total RNA from cells or tissues, isolation of tRNA, complete hydrolysis and dephosphorylation to generate individual modified and unmodified nucleosides. The LC-MS system is then used to quantitatively analyze and compare the abundance and proportions of each modification or unmodified nucleoside in the experimental and control groups.
Specifically, nucleosides with different masses and polarities have different retention times in the HPLC high-performance liquid chromatography, enabling the preliminary separation of different modified nucleosides. Subsequently, these nucleosides undergo ionization and enter the mass spectrometer. Peak areas are extracted for quantitative analysis of modified nucleosides based on the characteristic parent ion-to-fragment ion mass-to-charge ratio of each nucleoside and the retention time in HPLC.
Optimized pre-treatment and the use of high-quality Agilent equipment ensure comprehensive and accurate analysis of base modifications, elevating the sensitivity, precision, dynamic range, and robustness of the analysis results to a new level.
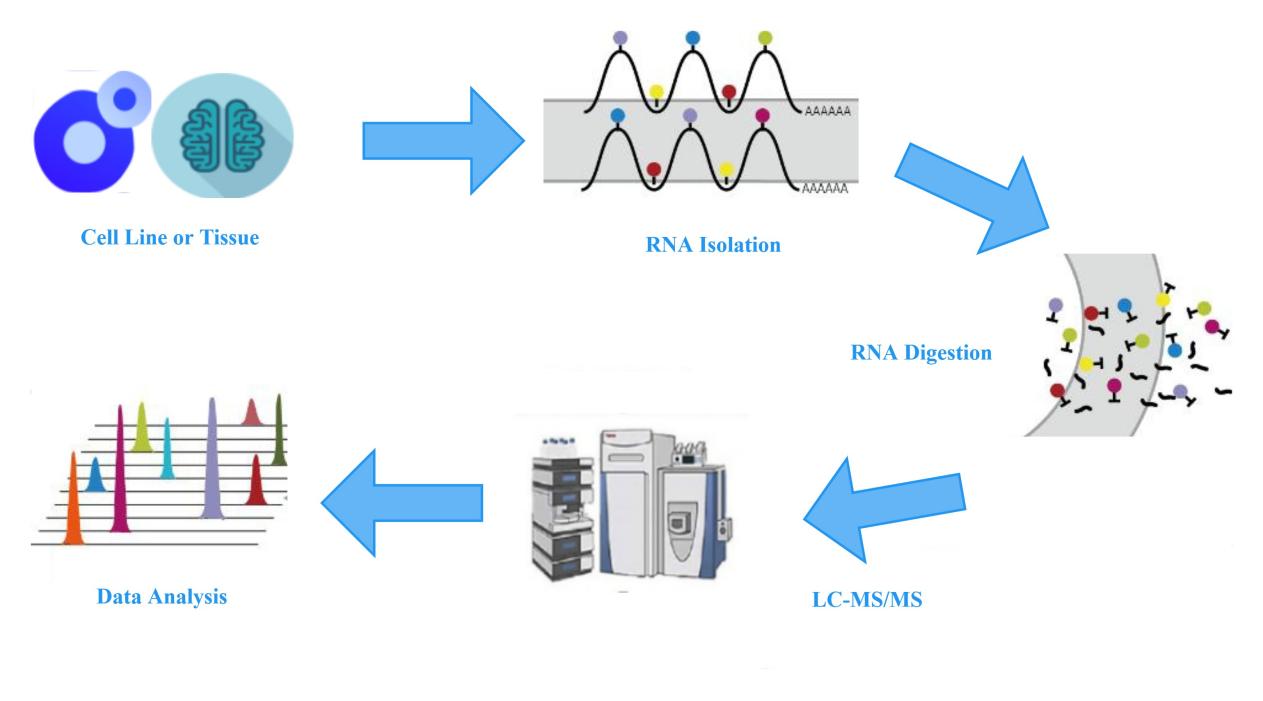 Figure 2: Experimental workflow for LC-MS measurement of tRNA modifications.
Figure 2: Experimental workflow for LC-MS measurement of tRNA modifications.
| Number | Nucleoside | Symbol | Number | Nucleoside | Symbol |
|---|---|---|---|---|---|
| 1 | 3′-O-methyladenosine | 3′-OMeA | 27 | 3'-O-methyluridine | 3'-OMeU |
| 2 | 2′-O-methylcytidine | Cm | 28 | 5-methyl-2-thiouridine | m5s2U |
| 3 | 3-methylcytidine | m3C | 29 | 5-methoxyuridine | mo5U |
| 4 | 5-methylcytidine | m5C | 30 | pseudouridine | Ψ |
| 5 | N6-isopentenyladenosine | i6A | 31 | 2'-O-methylinosine | Im |
| 6 | 5,2'-O-dimethylcytidine | m5Cm | 32 | 3-methyluridine | m3U |
| 7 | 1-methyladenosine | m1A | 33 | 1-methylpseudouridine | m1Ψ |
| 8 | 2-thiocytidine | s2C | 34 | 5-hydroxymethylcytidine | hm5C |
| 9 | N2,N2,7-trimethylguanosine | m2,2,7G | 35 | 5,2'-O-dimethyluridine | m5Um |
| 10 | N4-acetyl-2'-O-methylcytidine | ac4Cm | 36 | N6-threonylcarbamoyladenosine | t6a |
| 11 | N6-methyladenosine | m6A | 37 | 2-methylthio-N6-threonylcarbamoyladenosine | ms2t6A |
| 12 | 3'-O-methylcytidine | 3′-OMeC | 38 | 5-carboxymethyluridine | cm5U |
| 13 | 2'-O-methyladenosine | Am | 39 | 5-methoxycarbonylmethyl-2-thiouridine | mcm5s2U |
| 14 | N2, N2-dimethylguanosine | m22G | 40 | 5-Methoxycarbonylmethyluridine | mcm5U |
| 15 | 5'-O-methylthymidine | 5′-OMeT | 41 | 2-methylthio-N6-isopentenyladenosine | ms2i6A |
| 16 | 2′-O-methyluridine | Um | 42 | Peroxywybutosine | o2Yw |
| 17 | inosine | I | 43 | 5-taurinomethyl-2-thiouridine | tm5s2U |
| 18 | 2′-O-methylguanosine | Gm | 44 | 5-oxyacetic acid uridine | cmo5U |
| 19 | 1-methylguanosine | m1G | 45 | 5-carbamoylmethyuridine | ncm5U |
| 20 | 7-methylguanosine | m7G | 46 | Queuosine | Q |
| 21 | N2-methylguanosin | m2G | 47 | 5-taurinomethyluridine | tm5U |
| 22 | 3'-O-methylinosine | 3'-OMeI | 48 | 5-formyl-2′-O-methylcytidine | f5Cm |
| 23 | 2-thiouridine | s2U | 49 | dihydrouridine | D |
| 24 | 4-thiouridine | s4U | 50 | 5-formylcytidine | f5c |
| 25 | 5-methyluridine | m5U | 51 | wybutosine | yW |
| 26 | N4-acetylcytidine | ac4C | 52 | 5-methoxycarbonylmethyl-2'-o-methyluridine | mcm5Um |
Reference
Our products and services are for research use only.