
- Home
- PTMs Proteomics
- Proteomics Analysis of Lipoylation
Lipoylation, a pivotal post-translational modification (PTM), plays a critical role in diverse cellular processes. This modification involves the covalent attachment of lipoic acid to proteins, fostering structural and functional alterations. The impact of lipoylation extends across various biological pathways, underscoring its indispensability in cellular homeostasis.
In the field of cell biology, lipoylation is a multifaceted player that influences diverse cellular processes. Creative Proteomics follows the advances in analytical technology, coupled with a comprehensive exploration of its applications and complexities, in helping researchers delve into the dynamic landscape of lipoylation, new avenues for therapeutic intervention, and a deeper understanding of the cellular regulation it provides.
Glycine decarboxylase maintains mitochondrial protein lipoylation to support tumor growth
Journal: Cell Metab
Published: 2022
Background
The study focuses on understanding the role of glycine cleavage system (GCS) in hepatocellular carcinoma (HCC). While previous research has explored the importance of serine in supplying one-carbon units for nucleotide biosynthesis in cancer cells, the role of glycine oxidation, particularly through GCS, remains unclear. The study aims to investigate the significance of GCS flux in HCC and its potential impact on nucleotide biosynthesis, protein lipoylation, and mitochondrial activity.
Sample
The researchers examined hepatocellular carcinoma (HCC) cells, specifically HepG2, as well as other cell lines, to investigate the expression and metabolic flux of the glycine cleavage system. The study also involved mouse xenografts with HCC cells to explore the in vivo effects of glycine cleavage inhibition.
Technical Approach
The researchers employed a combination of stable and radioactive isotope tracing techniques along with computational flux decomposition to quantify mitochondrial glycine cleavage system (GCS) flux. They developed a novel method for measuring GCS flux using isotopically labeled serine and glycine and applied this approach to HCC cell lines. Additionally, they conducted genetic silencing experiments and utilized CRISPR-Cas9 knockout to investigate the effects of inhibiting glycine decarboxylase (GLDC), a key enzyme in GCS.
Results
The study revealed high GCS flux in hepatocellular carcinoma (HCC), supporting nucleotide biosynthesis(Figure 1). Surprisingly, GCS was found to play a crucial role in maintaining protein lipoylation and mitochondrial activity in addition to supplying one-carbon units. Genetic silencing of glycine decarboxylase inhibited lipoylation and impaired tumor growth, suggesting GLDC as a potential novel drug target for HCC. The research also highlighted the importance of considering the tissue of origin in understanding tumor-specific metabolic rewiring.
To systematically identify tumors with high glycine cleavage system activity, the authors analyzed the expression levels of four genes encoding GCS subunits (GLDC, AMT, GCSH, and DLD) in The Cancer Genome Atlas (TCGA). Two subunits of the GCS, GLDC and AMT, were found to be significantly overexpressed in hepatocellular carcinoma cells (HCC) and renal carcinoma (Figure 1).
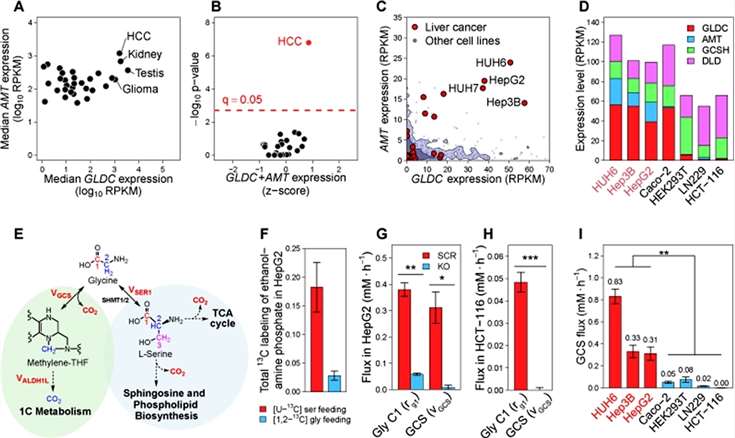 Figure 1
Figure 1
Treatment of HCC cells with glycine revealed an important contribution of GCS flux to endogenous purine and pyrimidine synthesis. The isotopic labeling fraction of folate-derived 1C units in ATP and deoxythymidine triphosphate (dTTP) was higher compared to non-HCC cell lines. In particular, in HepG2 cells, the authors found that approximately 7% of the 1C units in approximately ATP were derived from glycine, suggesting that GCS-derived 1C units flow to purine synthesis at a rate of 0.16 mM/h. Considering that GCS flux in HepG2 cells is 0.3 mM/h, the authors concluded that 1C units produced by GCS flux flow primarily to the purine synthesis pathway (Figure 2).
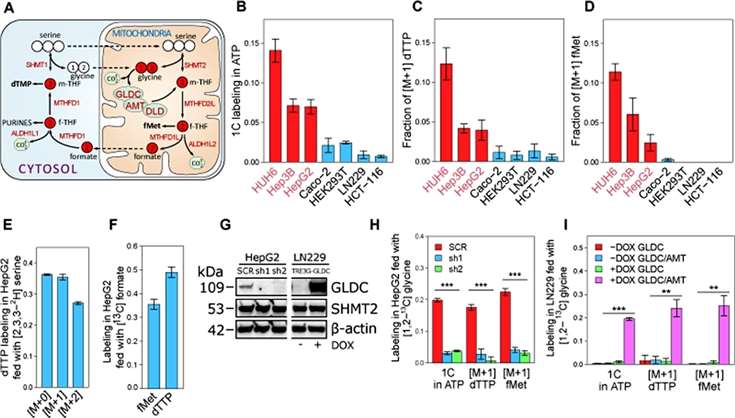 Figure 2
Figure 2
The results of the study showed that GLDC silencing inhibited the growth of HepG2, Hep3B, and HUH6 cells (Figure 3A), and inhibited the growth of tumors, with a 4-fold reduction in tumor size.
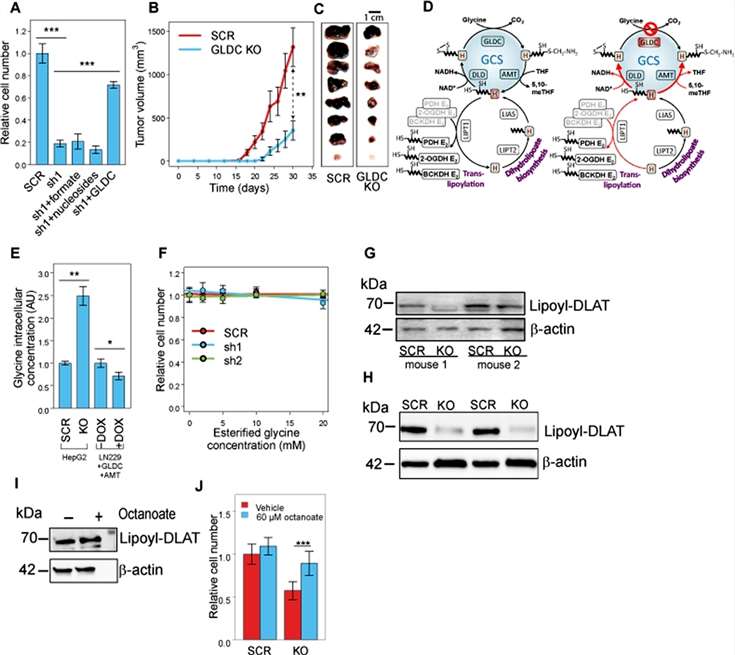 Figure 3
Figure 3
To investigate how GLDC-induced proteolipid acylation affects mitochondrial metabolism, the researchers examined the ability of mitochondria to oxidize a variety of metabolic substrates, and found that the ability to specifically oxidize pyruvate and α-KG, the lipoic acid-dependent complexes PDH and oxoglutarate dehydrogenase (OGDH), was markedly reduced in GLDC-silenced cells. In HepG2 GLDC-silenced cells, for both PDH and OGDH, a significant increase in the substrate-to-product ratio was further found (Figure 4).
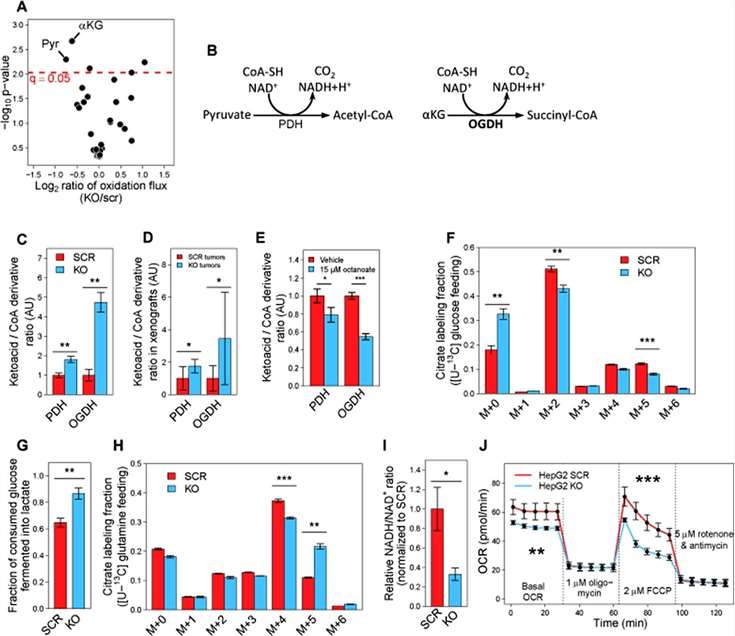 Figure 4
Figure 4
Conclusion
The study demonstrated the high expression of glycine cleavage system (GCS) genes and substantial glycine cleavage flux in hepatocellular carcinoma (HCC). While previous research has emphasized the role of glycine in cancer cell proliferation, this study revealed a novel connection between glycine cleavage and key cellular processes in HCC. The inhibition of glycine decarboxylase (GLDC), a crucial enzyme in GCS, resulted in the impairment of cell proliferation, both in vitro and in vivo, highlighting GLDC as a potential therapeutic target for HCC. The findings provided insights into the diverse roles of GCS, including its involvement in nucleotide biosynthesis, protein lipoylation, and mitochondrial metabolism. The study emphasized the importance of considering the tissue of origin in understanding metabolic rewiring in tumors and called for further exploration of GLDC as a potential drug target through the development of novel inhibitors and pre-clinical studies. Additionally, the researchers identified limitations, such as the need for further in vivo isotopic tracing experiments and investigation into the safety and potential resistance mechanisms associated with targeting GLDC in HCC.
Reference
Our products and services are for research use only.