
- Home
- PTMs Proteomics
- Proteomics Analysis of Kbz
Proteomics, the systematic study of proteins, plays a pivotal role in understanding cellular processes and their regulation. In recent years, a burgeoning interest has surrounded the proteomics analysis of Kbz, a compound of profound significance. Creative Proteomics is dedicated to protein modification research. Relying on our advanced technology platform and professional research team, we aim to unravel the intricate details of protein complementation within Kbz, revealing its biological significance and potential applications.
Kbz proteomics analysis is at the forefront of deciphering the intricate workings of this mysterious molecule. Through the application of advanced analytical techniques and the understanding of its PTM landscape, Creative proteomics can help researchers study the profound impact of Kbz on cellular activity. As technological advancest, proteomic analysis of Kbz promises to provide new discoveries with revolutionary consequences for biology, medicine, and beyond.
Lysine benzoylation is a histone mark regulated by SIRT2.
Journal: Nature Communications
Published: 2018
Background
The chromatin structure and transcriptional activity of genes are regulated by different protein post-translational modifications (PTM) in histones (or histone marks). There is growing evidence that the status of histone mark levels correlates with cellular metabolism. Benzoyl coenzyme A is a central intermediate in the degradation of a large number of aromatic growth substrates in bacteria and gut flora. In mammalian cells, one possible source of benzoyl coenzyme A is sodium benzoate (SB), one of the most commonly used preservatives in the world, with maximum allowable concentrations in food as high as 0.1%.SB has been successfully used as a drug for the treatment of patients suffering from acute hyperammonemia, a result of multiple urea cycle disorders. Although SB is classified by the FDA as a compound that is "generally recognized as safe," new research suggests that exposure to SB may cause harm to consumers. In addition, improperly dosing patients with intravenous SB can lead to serious complications. These findings suggest that human health risks are associated with SB, however, the underlying biological mechanisms remain unknown. In this paper, the authors report the histone marker lysine benzoylation (K bz ). This new histone mark was extensively characterized by using a variety of chemical and biochemical methods and detecting 22 K bz sites located on mammalian cellular histones.
Results
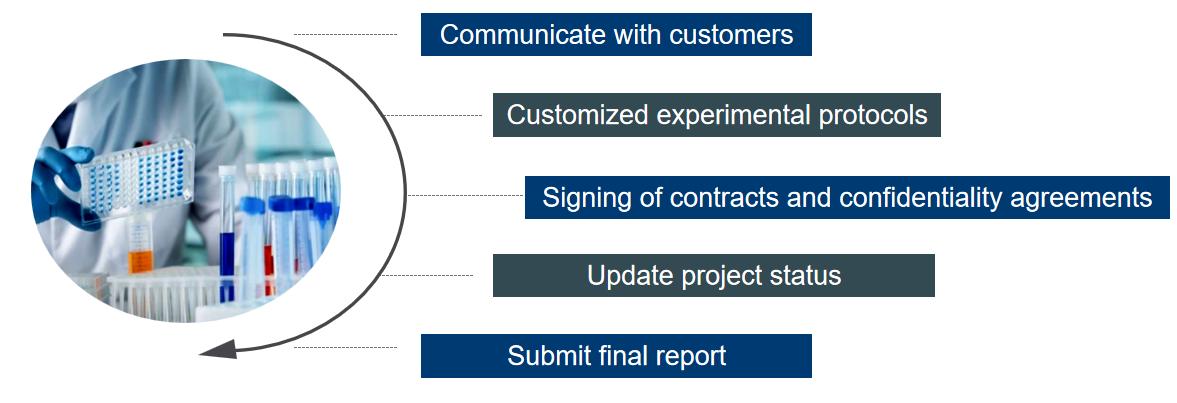
To discover new histone marks, the authors extracted core histones from HepG2 cells and subjected them to trypsin digestion. The resulting tryptic histone peptides were analyzed by high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS). The results showed that the MS/MS spectra of the two peptides matched very well and both peptides were co-eluted. In addition, similar results were obtained for two other peptides K +104.0261 STGGK ac APR and K +104.0260 QLATK ac AAR from histone H3 in HepG2 cells. Theoretically, only chemically identical peptides can show the same MS/MS fragmentation pattern and HPLC retention time. Therefore, our MS/MS and co-efflux analyses confirmed that the mass shift of +104.0268 Da was caused by the novel PTM K bz(Figure 1).
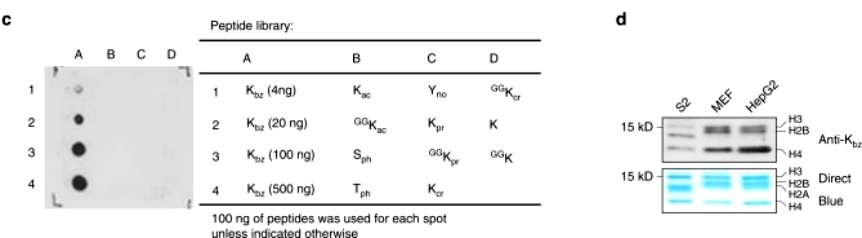 Figure 1
Figure 1
To identify histone K bz marks in vivo, the authors performed proteomic analysis using histones extracted from HepG2 and RAW cells treated with 5 mM SB. Extracted histones, with or without chemical propionylation, were digested and analyzed by trypsin. Spectra of identified K bz peptides were manually validated to eliminate false positives. Using these procedures and criteria, we identified 22 unique histone K bz sites. the K bz sites in HepG2 cells were distributed in a pattern similar to that in RAW cells. Interestingly, the 22 K bz sites in HepG2 or RAW cells were predominantly located in the N-terminal tail. In contrast, the histone K ac and K cr sites are more widely distributed, located in the N-terminal tail and other regions. This evidence suggests that K bz may have a different role in chromatin regulation than histones K ac and K cr.
To further test whether SB could be used by cells for Kbz, the authors extracted histones from D 5 -SB-treated HepG2 cells and chemically propionylated aliquots of the histones. The chemically propionylated and nonpropionylated histones were then trypsin digested and analyzed. Fifteen histone K bz sites with D5 were detected, further suggesting that SB is a precursor for benzoyl coenzyme A synthesis in the lysine benzoylation reaction(Figure 2).
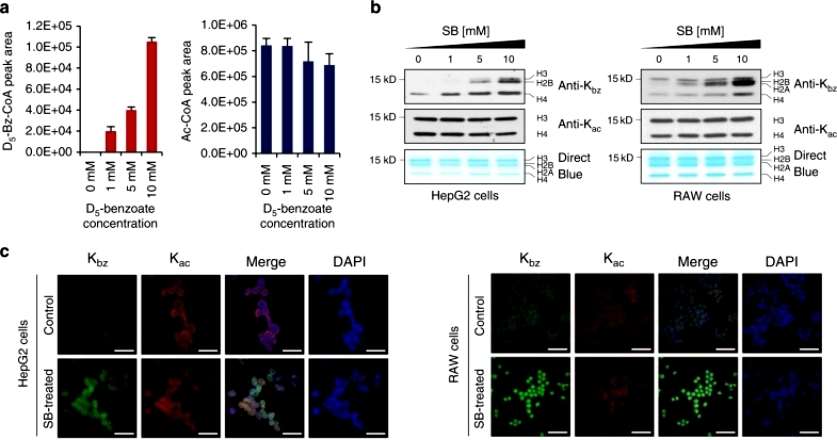 Figure 2
Figure 2
To confirm the direct debenzoylation activity of SIRT2, we performed in vitro debenzoylation reactions using two other synthetic peptides with benzoylated lysine residues (K bz STGGK ac APR and K bz QLATK ac AAR) as substrates. In both cases, we detected their corresponding unmodified counterparts. In contrast, no dephthaloylated peptide was detected in the absence of NAD +, which is an essential cofactor for the reaction. Furthermore, the reaction was inhibited by the class III HDAC inhibitor nicotinamide but not by TSA. These results suggest that SIRT2 is a debenzoylase in vitro(Figure 3).
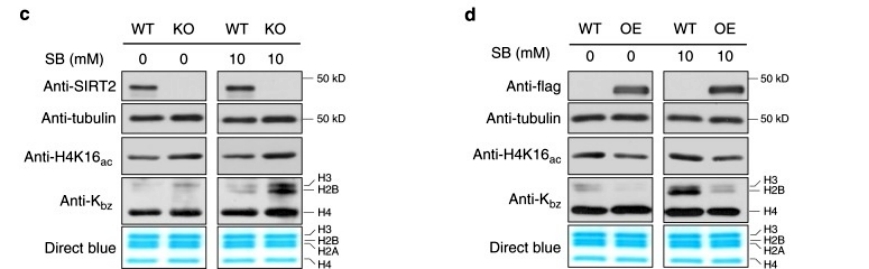 Figure 3
Figure 3
To investigate the genome-wide distribution of histone K bz marks, the authors performed ChIP-seq in HepG2 cells with a pan-anti-K bz antibody and also included a pan-anti-K ac antibody as a control. 20,283 K bz peaks distributed across 13,255 genes were identified in SB-treated HepG2 cells. Analysis of the ChIP-seq dataset revealed that more than half (57.33%) of the K bz signals were associated with gene promoters. Similar to K ac, K bz shows depletion in the TSS but is enriched around the TSS region with slightly higher signal upstream compared to downstream. This evidence suggests that histone K bz may be associated with transcription.
The authors examined the association between histone K bz marks and gene expression. To do this, we categorized all genes into four groups based on their expression levels and plotted the ChIP-seq peak signals of K bz on these genes. We observed a positive correlation between K bz levels and gene expression in TSS, which strongly supports a role for histone K bz in gene expression(Figure 4).
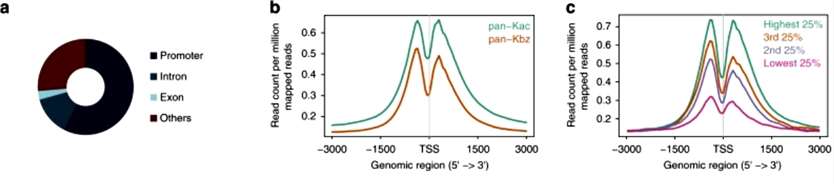 Figure 4
Figure 4
Conclusion
This experiment revealed that the histone lysine acylation response has physiological relevance and contributes to the regulation of chromatin structure and gene expression. Kbz has a larger molecular size and stronger hydrophobicity than other histones with short-chain fatty acids. In eukaryotes, benzoyl coenzyme A can be dose-dependently elevated with sodium benzoate (SB) concentration, which in turn increases histone benzoylation levels.SIRT2 is the only HDAC that removes Kbz in vitro.ChIP-seq and RNA-seq analyses revealed that histone Kbz labeling is involved in glycerophosphate metabolism, ovarian steroidogenesis, and phospholipase D signaling pathway-related expression of specific genes. The authors demonstrated that 5 mM SB (-0.07%) significantly increased histone Kbz levels at a concentration below the maximum allowable percentage allowed in food (0.1%), so this could raise potential safety concerns. Our study not only identified epigenetic and PTM pathways but also revealed potential mechanisms of physiological changes induced by SB.
Reference
Our products and services are for research use only.