
- Home
- PTMs Proteomics
- Proteomics Analysis of Iodination
Iodination, a crucial chemical process in the realm of biological studies, involves the incorporation of iodine atoms into molecules. This technique holds paramount importance in various fields, particularly in elucidating intricate biological structures and functions. The strategic introduction of iodine offers a powerful means to trace and analyze biomolecules, enabling researchers to delve deep into the intricacies of biological systems.
Iodination analysis stands as a cornerstone in the toolkit of biological researchers. The ability to selectively introduce iodine atoms into biomolecules empowers scientists to unravel the intricacies of biological systems at the molecular level. From understanding protein structures to advancing medical imaging techniques, iodination plays a pivotal role in advancing our knowledge of life sciences. The precision and versatility offered by iodination analysis underscore its significance in driving scientific discoveries and innovations.
Quantifying In Situ Structural Stabilities of Human Blood Plasma Proteins Using a Novel Iodination Protein Stability Assay
Journal: J Proteome Res
Published: 2022
Background
Many of the diseases that plague society today are driven by a loss of protein quality. One method to quantify protein quality is to measure the protein folding stability (PFS). Here, we present a novel mass spectrometry (MS)-based approach for PFS measurement, iodination protein stability assay (IPSA). IPSA quantifies the PFS by tracking the surface-accessibility differences of tyrosine, histidine, methionine, and cysteine under denaturing conditions. Relative to current methods, IPSA increases protein coverage and granularity to track the PFS changes of a protein along its sequence.
Results
The study applied the iodination protein stability assay (IPSA) to denatured serum as a control experiment, hypothesizing that this pre-denatured control would contain labeled sites but not generate "real" C1/2 values for those sites. Comparing pre-denatured serum to regular serum, it was observed that 66% fewer C1/2 sites passed quality filters in the pre-denatured serum. The study found that the quality control filters effectively identified quantifiable changes and removed flat or noisy denaturation curves. The control experiments confirmed that IPSA does not induce nonspecific damage to proteins and requires a folded protein structure for accurate measurements.
Regarding efficiency, the study compared IPSA with another method, the stability of proteins from rates of oxidation (SPROX), when applied to undepleted human serum. IPSA exhibited an overall label efficiency of 33.73% at the peptide level and 58.88% at the protein level, while SPROX showed label efficiencies of 31.41% at the peptide level and 72.54% at the protein level. The fitting efficiency of IPSA was 16.42% at the peptide level and 32.28% at the protein level, compared to SPROX with 4.34% at the peptide level and 14.86% at the protein level. Although SPROX had better efficiency in modifying methionine, IPSA provided double the protein coverage and four times the peptide coverage. IPSA measured more sites for proteins without methionine in the MS-detectable region and was deemed more suitable for quantifying PFS changes in multiple domains within the same proteins, as evidenced by the comparison of serum albumin, transferrin, and transthyretin (Figure 1).
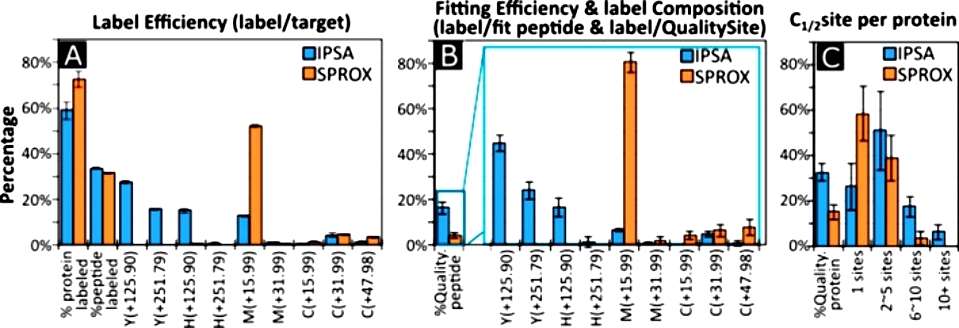 Figure 1
Figure 1
The study compared the iodination protein stability assay (IPSA) with the stability of proteins from rates of oxidation (SPROX) by examining the variation in C1/2 values between the two methods. Only four shared C1/2 sites were identified between the 283 sites from IPSA and the 45 sites from SPROX. However, when allowing proximal sites within 6 amino acid residues of each other, 30% of SPROX sites had a comparison in IPSA data, demonstrating high reproducibility (slope = 1.0021x, R2 = 0.93). The study emphasized that IPSA measures PFS reproducibly, similar to SPROX, but with a different distribution of reporter amino acids. Efficiency comparisons revealed that SPROX had better label efficiency in modifying methionine, but IPSA provided double the protein coverage and four times the peptide coverage. The study also assessed C1/2 variation between analytical approaches and found that IPSA data from QTOF-LFQ and Orbi-TMT runs were the most comparable and reproducible. Slope direction analysis demonstrated a highly reproducible slope direction from run to run, with QTOF-LFQ being the most reproducible among the tested methods (Figure 2).
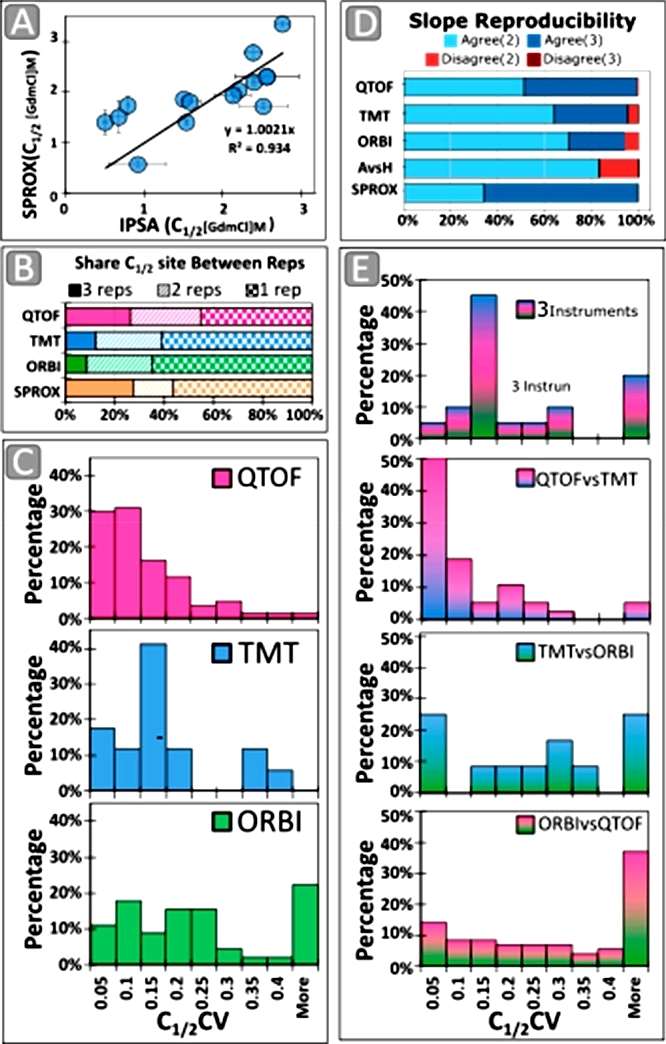 Figure 2
Figure 2
The study investigated whether the amino acid hydrophobicity correlated with the slope direction in the iodination protein stability assay (IPSA). A pronounced trend was observed, showing an increased frequency of negative slopes for histidine (H) relative to cysteine (C). Hydrophobicity, decreasing from C, methionine (M), and tyrosine (Y), to H, influenced the surface accessibility in folded structures. To further explore this, the study calculated the solvent-accessible surface area (SASA) values from published X-ray crystallography structures for proteins observed in the serum run (Figure 3).
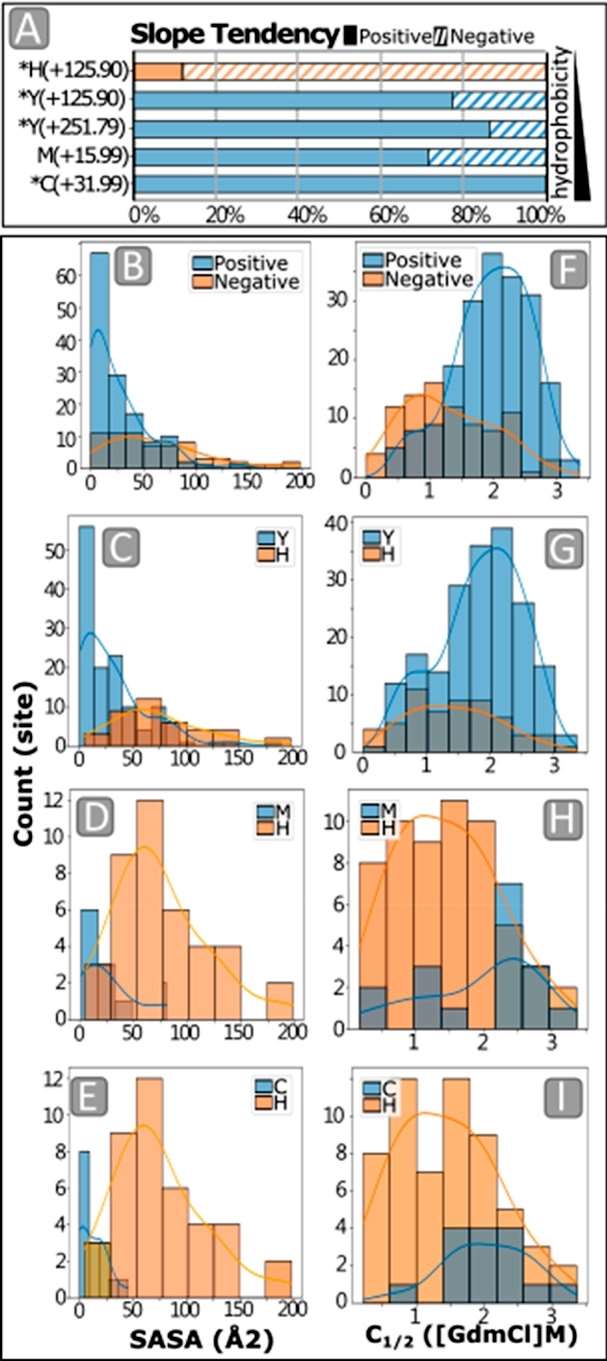 Figure 3
Figure 3
The study investigated changes in the structural stability of transferrin (TF) with the binding of iron using the iodination protein stability assay (IPSA). The study aimed to assess whether IPSA could detect C1/2 changes between different conditions and across the protein sequence. Purified apoTF and holoTF were subjected to IPSA, establishing a baseline. The study found that the C1/2 of Y115, an amino acid adjacent to the iron-binding site, was significantly higher in holoTF than in apoTF, indicating greater stability when both iron-binding sites are occupied. A comparison of site-specific C1/2 values across conditions showed that apoTF sites were generally lower than holoTF sites, and serum TF was slightly below holoTF. Shared site C1/2 values were statistically significant between apo and holo conditions. The results implied that as soon as one of the TF iron binding sites is occupied, the overall protein structure is stabilized, consistent with the observation that the two iron binding sites have different affinities. The study suggested that IPSA has the sensitivity to quantify PFS differences between protein states and that individual sites in the protein report independent values for conformational change. Overall, the findings supported the conclusion that IPSA can measure meaningful differences in PFS between different conditions for individual sites in the protein (Figure 4).
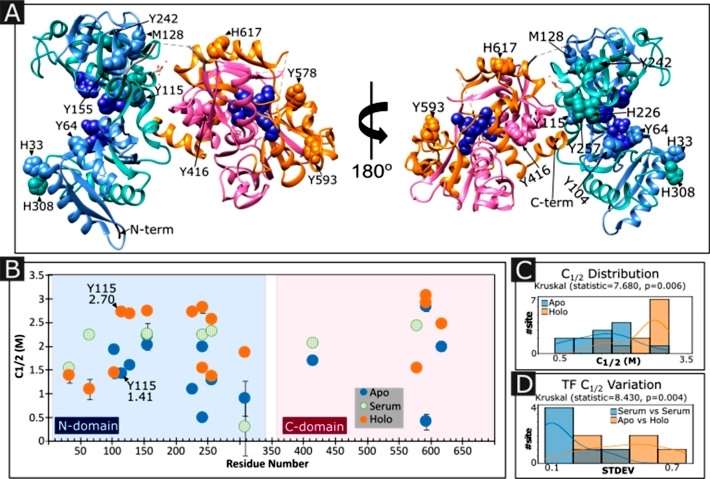 Figure 4
Figure 4
Conclusion
The article introduces a new mass spectrometry-based method, the iodination protein stability assay (IPSA), which utilizes irreversible iodination and oxidation to modify specific amino acids. IPSA increases proteome coverage and provides granularity in assessing residue-specific PFS changes within proteins. The study applies IPSA to human serum proteins, demonstrating its capability to reproducibly measure PFS in complex mixtures at near-physiological conditions. The article discusses the assay's efficiency, reproducibility, and potential in studying structural details, emphasizing its relevance in the context of the human serum proteome. The study compares the PFS of transferrin proteins in serum versus purified forms, highlighting the technique's ability to detect folding changes in situ. Overall, the article suggests that IPSA has the potential to advance understanding of protein folding changes in disease contexts and lays the foundation for future applications of this technique.
Reference
Our products and services are for research use only.