
- Home
- PTMs Proteomics
- Proteomics Analysis of Deamidation
Non-enzymatic catalyzed deamidation of asparagine (Asn) and glutamine (Gln) residues in proteins occurs from time to time in vivo or in vitro as a specific post-translational modification (PTM), which can affect protein structure, stability, folding and aggregation. In particular, deamidation of certain positions within monoclonal antibody molecules is known to affect the safety and efficacy of the product, and is therefore often regarded as a critical quality attribute (CQA). Meanwhile, protein deamidation plays an important role in aging, neurological diseases, age-related diseases and autoimmune diseases.
To comprehend the biological significance of deamidation, precise identification and measurement are crucial. When examining deamidation, various analytical techniques are used, including:
Deamidation, a multifaceted phenomenon in protein biology, warrants comprehensive investigation due to its implications in health, disease, and biotechnology. Creative Proteomics relies on advances analyse techniques continue to enhance scientists understand of deamidation, our services will offer valuable insights into its mechanisms and applications. As we delve deeper into the world of protein modifications, the study of deamidation emerges as a crucial frontier in understanding the intricate interplay between structure and function in biological systems.
N-Linked Glycosylation Prevents Deamidation of Glycopeptide and Glycoprotein
Journal: ACS Chem Biol
Published: 2020
Background
Deamidation is a spontaneous, non-enzymatic deamidation reaction of asparagine (Asn) and glutamine (Gln) in peptides or proteins, and is a common pathway of protein degradation. During the deamidation of Asn, a five-membered cyclosuccinimide intermediate is formed, which hydrolyzes in different directions to form aspartic acid (Asn) and β-Asp (isoAsp). During aging of peptides and proteins, the deamidation site switches from electrically neutral to negatively charged and forms a β-conjugation in the peptide backbone, leading to structural and functional disruption of the protein. Deamidation of physiological proteins has been implicated in several human diseases and is considered to be a "timer" of disease.
Results
The authors selected five peptides from the Uniprot database of glycoproteins as targets for their study. The sequences of peptides 1a and GP 1 (GlcNAcated peptides) were selected from human α-1-acid glycoprotein 1, the sequences of peptides 2a and GP 2 were selected from the peptidyl prolyl cis-trans-isomerase FKBP10, the sequences of peptides 3a, GP 3, and GP 6 were selected from the deoxyribonuclease-2-alpha, the sequences of peptides 4a and GP 4 were selected from the sidekick-1 protein, and the sequences of peptides 5a and sequences of GP 5 were selected from Smo protein homologs. The sequences of these peptides are NGT, NHT, NSS, NDS, and NAT, respectively (Figure 1).
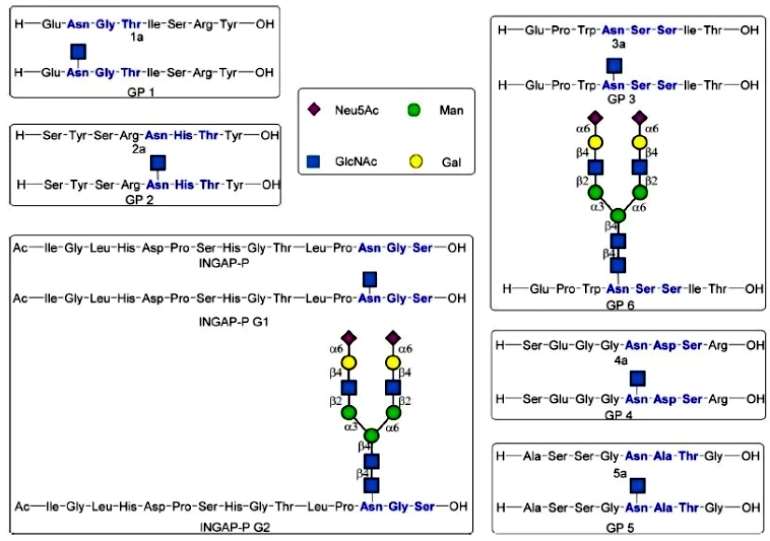 Figure 1
Figure 1
The NGT sequence has the smallest site resistance at the N+1 position and is the fastest deamidated. Only 32.6% of peptide 1a remained undeamidated after 48 h of incubation, and the main degradation product was a diketopiperazine-containing peptide obtained from the reaction of the N-terminal amino group with the succinimide intermediate. The half-life of the deamidation of peptide 1a was calculated to be 31 h; whereas after incubation of the corresponding glycopeptide GP 1 under the same conditions for 5 days, the main peak remained the original GP 1. The spatial site resistance at the N+1 position was large in the NHT, NSS, and NDS sequences, but the imidazole group of histidine, the hydroxyl group of serine, and the carboxylic acid group of aspartic acid were catalytic for the deamidation reaction. The half-lives of peptides 2a, 3a and 4a were 6.4 days, 13 days and 13.2 days, respectively. the main peak of HPLC of GP 2 was still the original peak of GP 2 after 5 days of incubation under the same conditions; no deamidation product was observed for GP 3 after up to 40 days of incubation; and GP 4 was not the product peak of deamidation, although new peaks were generated after 40 days of incubation. the NAT sequence has a greater spatial site resistance than the NGT The NAT sequence, with greater spatial site resistance and no catalytic effect of the methyl side chain, was the slowest to deamidate. The deamidation half-life of peptide 5a was 18.78 days, while GP 5 did not show any deamidation product after 20 days of incubation. All of these results demonstrate that glycosylation of peptides reduces their deamidation rate and makes them more stable. In addition, the authors demonstrated that the naturally N-glycosylated GP 6 has the same function of preventing the deamidation reaction as the mono-GlcNAc glycosylated GP 3 (Figure 2).
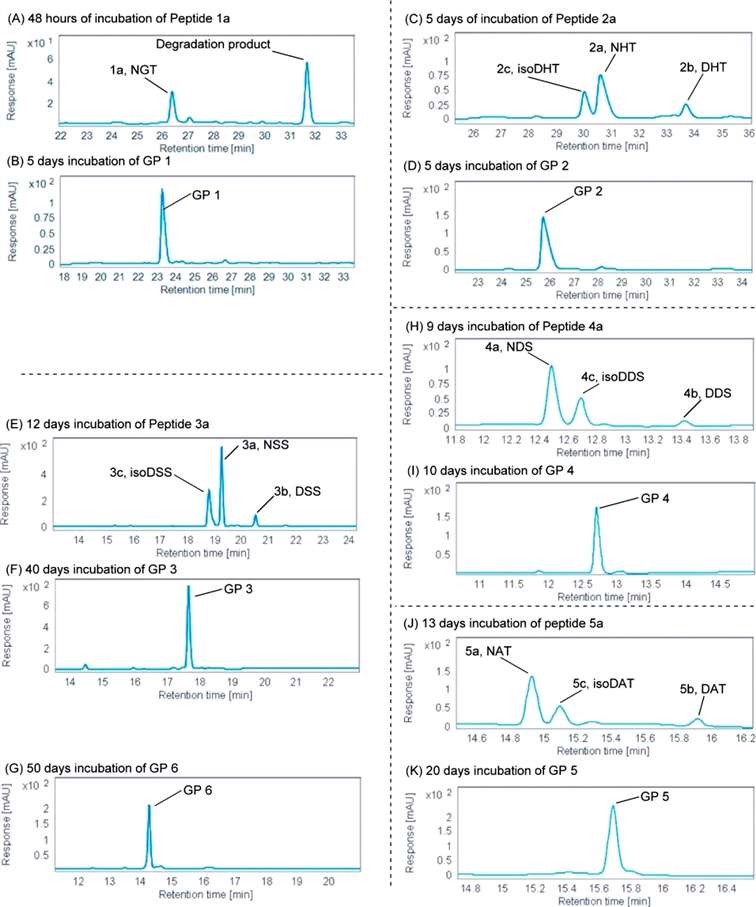 Figure 2
Figure 2
Subsequently, the authors explored the preventive effect of N-glycans on glycopeptide deamidation using the natural glycoprotein RNase B as a substrate.RNase B and RNase A contain the same amino acid sequence and have similar biological functions; the only difference between them is the high-mannose-type N-glycosylation on residue Asn34. The authors incubated RNase A in 0.1 M glycine-NaOH strong alkaline buffer (pH = 9.6) at 78°C for 4 days to allow the amide reaction to proceed faster, and mass spectrometry analysis was performed on the cleaved fragment NLTK after trypsin digestion. The results showed that the majority of the Asn34 residue of RNase A was deamidated and the NLTK was converted to DLTK and IsoDLTK, whereas RNase B was incubated for 4 days under the same conditions, and the high mannose-type N-glycans other than the first GlcNAc were removed by Endo H. After digestion and mass spectrometry analysis, the main peak was still GlcNAcated Asn34, and there was no deamidation peak. and no deamidation peak.
The authors further applied the N-glycosylation strategy to stabilize the therapeutic peptide INGAP-P, which has been shown to be therapeutic for both type 1 and type 2 diabetes.The presence of the NGS sequence at the C-terminus of INGAP-P resulted in only 60% of the peptide being present as non-deamidated after 40 hours of incubation, with a calculated half-life of 55.9 hours. In contrast, both GlcNAc-glycosylated INGAP-P (INGAP-P G1) and N-glycosylated INGAP-P (INGAP-P G2) were more stable than unmodified INGAP-P, and after 30 days of incubation, the HPLC result was only a single peak of the original (Figure 3).
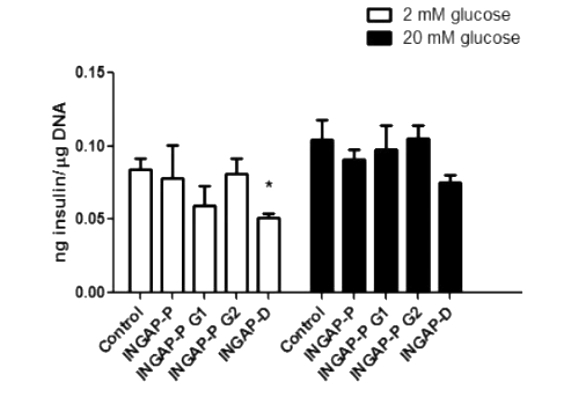 Figure 3
Figure 3
Conclusion
Since it was shown that INGAP peptide can stimulate insulin secretion from pancreatic β-cells, the authors further explored the effect of modified INGAP peptide on insulin secretion in the hamster insulinoma cell line HIT-T15. However, the results showed that there was no significant difference in insulin secretion between control, unmodified INGAP-P peptide, and glycosylated INGAP-P peptide at basal levels and glucose-stimulated conditions. In contrast, a certain reduction in insulin secretion was observed in cells treated with deamidated INGAP-P (INGAP-D) (Figure 7), which may be due to its reduced biological activity. In this paper, we demonstrated that N-glycosylation in glycoproteins and glycopeptides can reduce the deamidation rate to a low level, but N-glycosylation does not protect the proteins from other chemical degradation when the incubation time is prolonged up to a certain time. This strategy is instructive for the development of new biotechnologies to increase the stability of peptide and protein drugs.
Reference
Our products and services are for research use only.