
- Home
- PTMs Proteomics
- Proteomics Analysis of Benzoylation
Proteomics, the comprehensive study of proteins in biological systems, plays a pivotal role in deciphering intricate cellular processes. Among the myriad post-translational modifications (PTMs), benzoylation has emerged as a significant focus, offering insights into protein function, regulation, and signaling pathways.
The proteomics analysis of benzoylation represents a frontier in deciphering the complex language of cellular regulation. From unraveling molecular mechanisms to exploring therapeutic avenues, the study of benzoylation in PTMs has far-reaching implications. As analytical techniques continue to evolve, Creative Proteomics's ability to dissect the role of benzoylation in health and disease will undoubtedly expand, opening new vistas in the field of proteomics.
Lysine benzoylation is a histone mark regulated by SIRT2.
Journal: Nature Communications
Published: 2018
Background
The chromatin structure and transcriptional activity of genes are regulated by different protein post-translational modifications (PTM) in histones (or histone marks). There is growing evidence that the status of histone mark levels correlates with cellular metabolism. For example, short-chain fatty acids can be produced by cellular metabolism or derived from fermentation by the gut microbiota. These metabolites can be used as precursors for the generation of metabolites that can be used for histone acylation Some examples of these metabolites include crotonic acid, butyric acid, alpha-ketoglutaric acid, and 3-hydroxybutyric acid. The presence of multiple different acyl-coenzyme A's adds an interesting possibility that cellular metabolism influences epigenetics through metabolite-directed histone labeling of undescribed pathways.
Benzoyl coenzyme A is a central intermediate in the degradation of a large number of aromatic growth substrates in bacteria and gut flora. In mammalian cells, one possible source of benzoyl coenzyme A is sodium benzoate (SB), one of the most commonly used preservatives in the world, with maximum allowable concentrations in food as high as 0.1%.SB has been successfully used as a drug for the treatment of patients suffering from acute hyperammonemia, a result of multiple urea cycle disorders. Although SB is classified by the FDA as a compound that is "generally recognized as safe," new research suggests that exposure to SB may cause harm to consumers. In addition, improperly dosing patients with intravenous SB can lead to serious complications. These findings suggest that human health risks are associated with SB; however, the underlying biological mechanisms remain unknown.
Here, the authors report the histone marker lysine benzoylation (K bz ). The authors extensively characterized this new histone mark using a variety of chemical and biochemical methods and assayed 22 K bz sites located on mammalian cellular histones. Metabolic labeling experiments showed that SB can stimulate histone K bz by producing cytosolic benzoyl CoA. Importantly, ChIP-seq (chromatin immunoprecipitation followed by sequencing) and RNA-seq results indicate that histone K bz is a gene promoter-rich mark with unique physiological relevance. We further showed that SIRT2, a NAD+-dependent protein deacetylase, can remove Kbz in vitro and in vivo. Thus, the present study identifies an epigenetic mechanism that may lead to physiological changes induced by widely used food preservatives.
Results
To discover new histone marks, the investigators extracted core histones from HepG2 cells and subjected them to trypsin digestion. The resulting trypticized histone peptides were analyzed by high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS); the resulting MS/MS data were analyzed by PTMap software using an unrestricted sequence comparison algorithm that searches for mass shifts caused by PTM. Notably, histone H2B peptide PEPTK +104.0268 SAPAPK has a mass shift of +104.0268 Da at the lysine residue position H2BK5. This precise mass shift was used to deduce the possible elemental composition with a maximum allowable amount of 2 nitrogen atoms and a mass tolerance of ±0.02 Da . The results showed that the MS/MS spectra of the two peptides matched very well and both peptides were co-eluted. In addition, similar results were obtained for two other peptides K +104.0261 STGGK ac APR and K +104.0260 QLATK ac AAR from histone H3 in HepG2 cells. Theoretically, only chemically identical peptides can show the same MS/MS fragmentation pattern and HPLC retention time. Therefore, our MS/MS and co-efflux analyses confirmed that the mass shift of +104.0268 Da was caused by the novel PTM K bz (Figure 1).
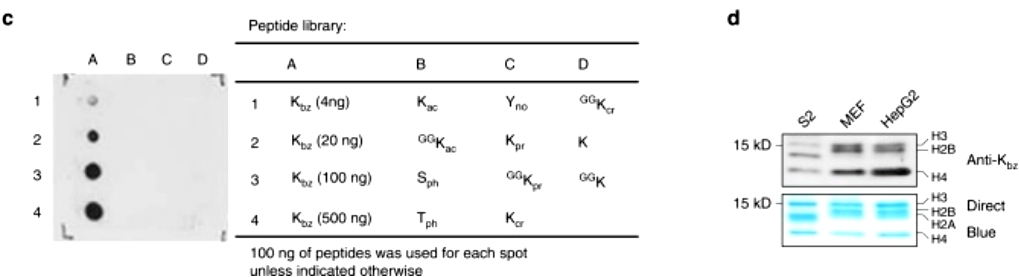 Figure 1
Figure 1
To identify histone K bz marks in vivo, we performed proteomic analysis using histones extracted from HepG2 and RAW cells treated with 5 mM SB. Extracted histones, with or without chemical propionylation, were digested and analyzed by trypsin. Spectra of identified K bz peptides were manually validated to eliminate false positives. Using these procedures and criteria, we identified 22 unique histone K bz sites.The K bz sites in HepG2 cells were distributed in a pattern similar to that in RAW cells. Interestingly, the 22 K bz sites in HepG2 or RAW cells were predominantly located in the N-terminal tail. In contrast, the histone K ac and K cr sites are more widely distributed, located in the N-terminal tail and other regions. This evidence suggests that K bz may have a different role in chromatin regulation than histones K ac and K cr (Figure 2).
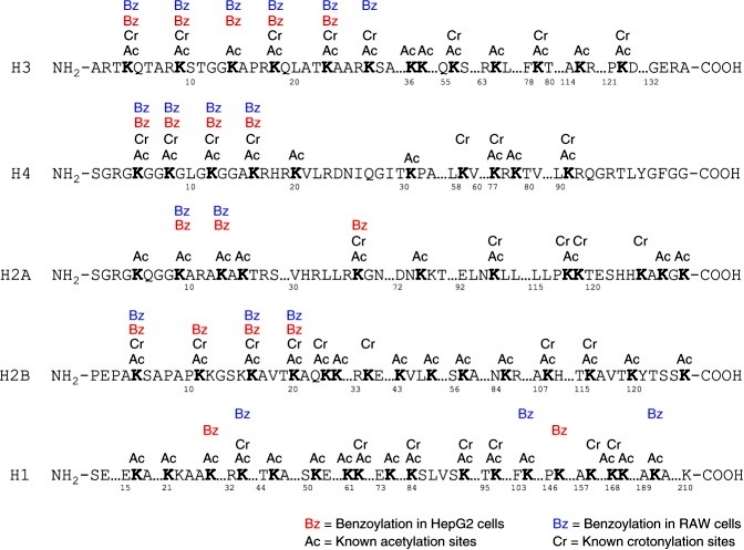 Figure 2
Figure 2
To further test whether SB can be used by cells for Kbz, we extracted histones from D 5 -SB-treated HepG2 cells and chemically propionylated aliquots of histones. The chemically propionylated and nonpropionylated histones were then trypsin digested and analyzed. Our experiments detected 15 histone K bz sites with D5, further suggesting that SB is a precursor for benzoyl coenzyme A synthesis in the lysine benzoylation reaction(Figure 3).
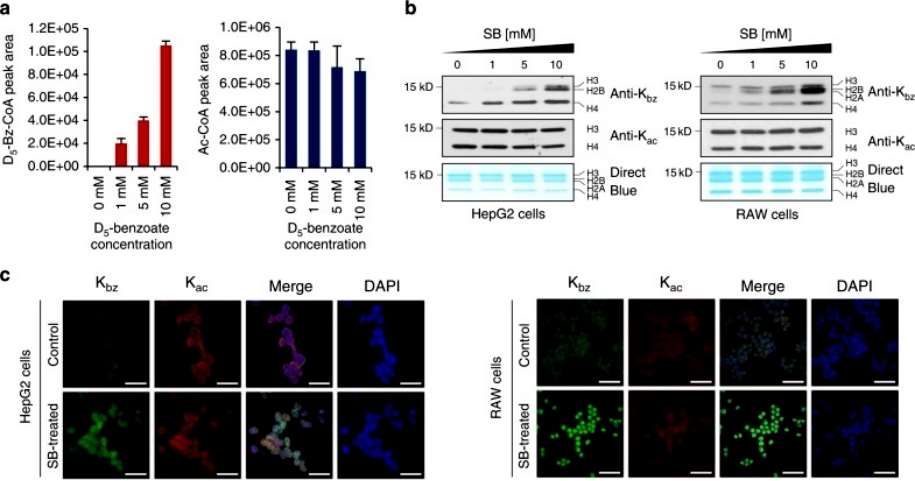 Figure 3
Figure 3
Conclusion
The histone lysine acylation reaction has physiological relevance and contributes to the regulation of chromatin structure and gene expression. Kbz has a larger molecular size and stronger hydrophobicity than other histones with short-chain fatty acids. In eukaryotes, benzoyl coenzyme A can be dose-dependently elevated with sodium benzoate (SB) concentration, which in turn increases histone benzoylation levels. SIRT2 is the only HDAC that removes Kbz in vitro.ChIP-seq and RNA-seq analyses revealed that histone Kbz labeling is involved in glycerophosphate metabolism, ovarian steroidogenesis, and phospholipase D signaling pathway-related expression of specific genes. The authors demonstrated that 5 mM SB (-0.07%) significantly increased histone Kbz levels at a concentration below the maximum allowable percentage allowed in food (0.1%), so this could raise potential safety concerns. Our study not only identified epigenetic and PTM pathways but also revealed potential mechanisms of physiological changes induced by SB.
Reference
Our products and services are for research use only.