
- Home
- PTMs Proteomics
- Protein PTM Qualitative Analysis
Proteins undergo significant changes in physicochemical properties after post-translational modifications (PTMs), resulting in exponential amplification of protein function. Due to the numerous potential modification sites on proteins, precise localization of the modifications is a prerequisite for studying the modifications' regulation of protein function. Currently, mass spectrometry (MS) is the main method for protein PTM research. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) based analysis can accurately characterize the molecular mass of a protein or peptide and can detect which modifications have occurred at the site of the protein or peptide. Creative Proteomics is a comprehensive contract research organization (CRO) with extensive experience in PTM identification. We are committed to providing precise localization and prediction of PTM sites to meet our client's specific project needs, including phosphorylation, glycosylation, acetylation, ubiquitination, propionylation, butyrylation, malonylation, and more.
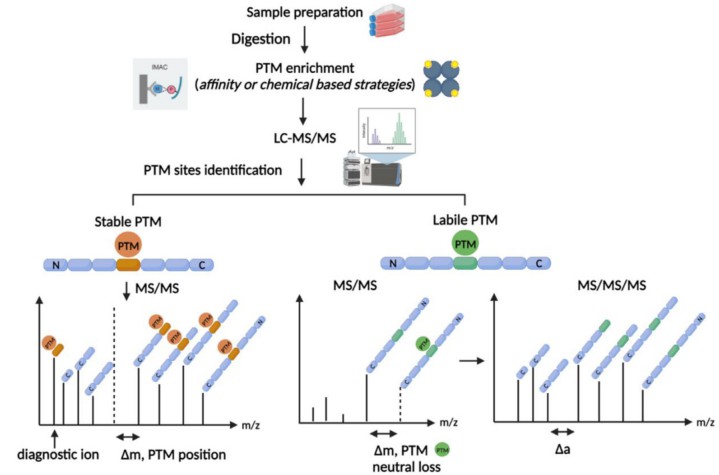 Fig. 1 MS/MS fragmentation of PTMs. (Brandi, Jessica, et al, 2022)
Fig. 1 MS/MS fragmentation of PTMs. (Brandi, Jessica, et al, 2022)
| Some common PTMs and their target amino acids, mass shift, and main functions | |||
|---|---|---|---|
| PTM types | Target amino acid residues | Mass shift/Gross formula shift (stable in MS/MS fragmentation) | Main biological functions |
| Phosphorylation | Ser, Thr, Tyr, His, Arg, Lys, Asp, Cys, Glu | +79.9663 (HPO3) | Protein activity, stability, cellular location, signalling pathways |
| Acetylation | Lys, Ser, Thr, N terminus | +42.0105 Da (CH3CO) | Protein folding, epigenetics, gene transcription, DNA damage repair, cell division, signal transduction, autophagy and metabolism |
| Ubiquitination | Lys, N terminus, Cys, Ser, Thr | +114.043 Da (Gly-Gly) | Protein degradation, trafficking, regulation of enzymes, translation, DNA repair |
| Glycosylation | Asn, Arg (N-linked) Ser, Thr, Tyr (O-linked) | >800 Da +203.0793 Da (HexNAc) +291.095 Da (Sialic acids) +365.148 Da (HexHexNAc) +162.0528 Da (Hexose) | Protein function, structure, stability, cellular adhesion, enzyme activity, protein trafficking |
| Methylation | Lys, Arg, His, Ala, Asn | +14.01565 Da (CH3) +28.0313 Da (C2H6) +42.04695 Da (C3H9) | Activity of specific proteins, epigenetics, cell cycle progression |
| Sumoylation | Lys | 600.2503 (SUMO-1) 599.2663 (SUMO-2/3) | Protein interactions, subcellular localization, enzymatic activities |
Based on effective protein extraction methods, peptide separation and enrichment strategies, and advanced mass spectrometers, Creative Proteomics is dedicated to providing protein PTM identification services, including identification of a targeted PTM and the global profiling of PTMs within the proteome.

Creative Proteomics strives to be a guide and assistant in protein PTM research for our customers worldwide. For more information about our PTM identification services or if you have any questions about our services, please contact us. We will provide you with strong technical support to save your heart and effort.
References
Our products and services are for research use only.