
Creative Proteomics is a leading custom service provider in post-translational modification (PTM) proteomics analysis. Our expert team has many years of experience in the bioinformatics analysis of protein PTMs. We currently provide protein PTM bioinformatics analysis services which include PTM site prediction service, PTM site motif analysis service, PTM crosstalk prediction service, and PTM functional analysis service. With a team of experts and state-of-the-art equipment, our PTM proteomics analysis platform helps our clients navigate successful research projects through robust workflows, robust analysis, and tailored services.
PTMs are biochemical reactions that occur after protein translation and alter the regulatory physicochemical properties, maturation, and activity of most proteins through cleavage, folding, ligand binding, and the addition of modifying groups to one or more amino acids. To date, more than 600 types of PTMs have been identified. They have been shown to have a range of cellular functions, and each modification has multiple effects on protein structure and function involving protein signaling, activity, stability, metabolism, and localization. PTMs also play key roles in a wide range of biological regulatory mechanisms, such as signaling pathways, protein-protein interactions, DNA damage response, and metabolic pathways. Therefore, the dysregulation in PTMs can lead to a variety of diseases, including diabetes, cardiovascular disease, neurological disorders, and different cancers.
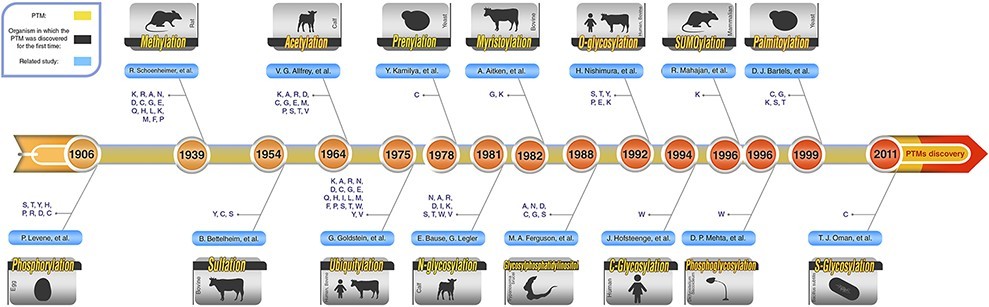 Fig.1 Schematic PTM discovery timeline for 10 major PTM. (Ramazi, Shahin, and Javad Zahiri., 2021)
Fig.1 Schematic PTM discovery timeline for 10 major PTM. (Ramazi, Shahin, and Javad Zahiri., 2021)
The prediction of protein PTMs is currently an important research topic in the field of protein bioinformatics. PTMs allow proteins to adopt more complex structures, have more sophisticated functions, and perform different tasks more precisely. Based on our experienced experts and multiple machine learning algorithms, we are able to provide not only common PTM site prediction services, such as phosphorylation site, glycosylation site, acetylation site, methylation site, SUMOylation site and ubiquitination site prediction, but also new PTM site prediction services.
Motif analysis can provide useful sequence pattern information for all identified PTM proteins/peptides and help identify important motifs. On the other hand, based on known substrate specificity, motif analysis can ideally help us identify relevant enzymes and thus further understand the biological significance involved in each biological process.
Proteins can be modified at multiple sites as well as by multiple types of PTMs. The combinatorial action of multiple PTMs on the same or on different proteins to obtain higher-order regulation is called "crosstalk". The study of PTM crosstalk is highly relevant to trigger their role in biological processes. It is necessary to identify the biological context/networks in which PTM event functions combined to cross-talks relate to physiological or pathological states by computational prediction. To meet the increasing demand for PTM crosstalk analysis, we are now offering a PTM crosstalk prediction service.
Functional analysis aims to bring the quantitative PTM analysis into the biological context and interpret data at the biological level based on annotations, protein subcellular localization, protein structural properties, protein interactions, or metabolic fluxes. Our service is based on a variety of popular functional analysis tools, enabling fast and targeted research while still providing an in-depth overview of the potential role of PTMs in cellular processes.
To simplify the analysis of complex PTM data and to improve our clients' understanding of various PTMs in different organisms, Creative Proteomics offers PTM bioinformatics analysis services. Our bioinformatics team has done a lot of work on the identification of PTM results and has developed our own platform for quality control, data processing, and analysis. Contact us for all the details!
Reference
Our products and services are for research use only.