Introduction to GlycoRNA Sequencing
In the rapidly evolving field of epitranscriptomics, GlycoRNA sequencing has emerged as a powerful new tool for decoding a previously overlooked layer of RNA biology. Glycosylated RNAs (glycoRNAs)—RNAs that carry covalently attached glycan structures—represent a novel class of post-transcriptionally modified molecules that bridge RNA biology and glycoscience. The recent discovery of glycoRNAs on the cell surface has disrupted long-held paradigms and opened the door to a new frontier in RNA research.
Unlike traditional RNA modifications, RNA glycosylation was once thought implausible due to the lack of known glycosylation pathways in RNA biology. However, landmark studies have now demonstrated that small noncoding RNAs can indeed be glycosylated and trafficked to the cell membrane, where they may engage in cell–cell communication and immune recognition. These discoveries have sparked intense interest across biomedical research and pharmaceutical development communities.
Despite its transformative potential, glycoRNA detection presents significant analytical challenges due to the structural complexity and low abundance of glycosylated RNA species. Conventional RNA sequencing methods are not designed to identify or quantify these modifications. This gap has fueled the need for specialized GlycoRNA sequencing services that combine targeted enrichment, advanced sequencing platforms, and glycan-specific data interpretation.
Biological Significance of GlycoRNAs
Structured and Selective GlycoRNA Expression
Emerging research shows that glycoRNAs aren’t random by-products. Instead, they appear to be:
- Structurally defined: Enriched in small non-coding RNAs like Y RNAs and small nuclear RNAs.
- Selectively expressed: Their patterns vary by cell type, developmental stage, and physiological conditions.
- Immunologically relevant: For example, glycoRNAs can bind Siglecs—a group of immune receptors that play key roles in immune cell signalling.
Linking Trafficking Pathways to Post-Transcriptional Control
Current findings suggest a tight interplay between:
- RNA trafficking mechanisms that export these modified RNAs to the membrane.
- Post-transcriptional regulation, which governs which RNAs get glycosylated and when.
A New Axis Between the Transcriptome and the Glycome
From a systems biology standpoint, glycoRNAs may act as a bridge between two major molecular landscapes—the transcriptome (all RNAs in a cell) and the glycome (all glycan structures). This crosstalk opens up:
- Novel drug discovery targets, particularly in immune-oncology and infectious diseases.
- Fresh biomarkers for diseases where glycan signalling is disrupted.
- New therapeutic windows for modifying cellular responses or improving biologics efficacy.
Overview of GlycoRNA Sequencing Technology
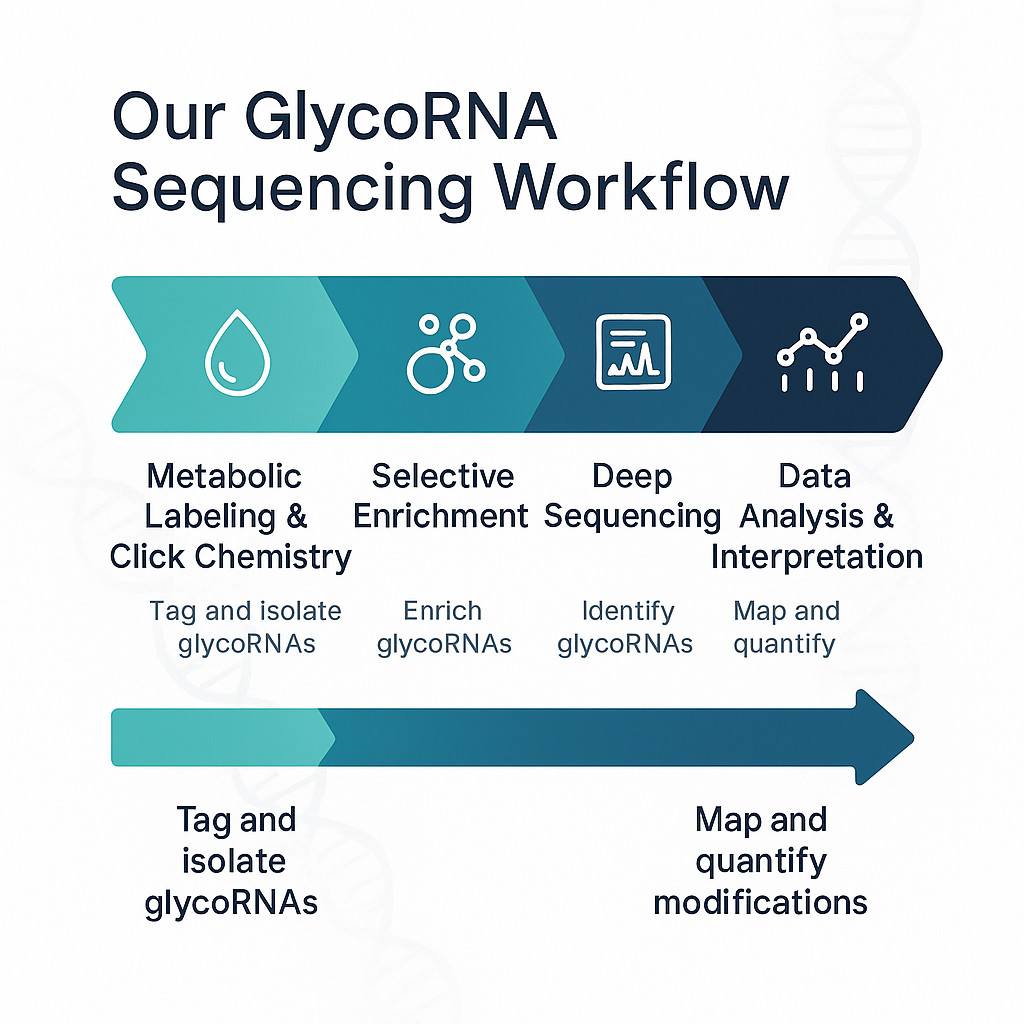
Detecting glycoRNAs requires specialized techniques that integrate glycan labeling, RNA enrichment, and high-resolution sequencing. Conventional RNA-seq alone is ill-suited for identifying glycosylated RNA species, making GlycoRNA sequencing technology indispensable for accurate analysis.
By combining metabolic labeling, tailored library preparation, deep sequencing, specialized mass-spectrometry, spatial imaging, and advanced computational analysis, GlycoRNA sequencing robustly detects and characterizes glycosylated RNAs—unlocking a new sensory dimension in RNA biology research.
1. Metabolic Labeling & Chemical Tagging
Ac₄ManNAz incorporation: Live cells are cultured with azide-tagged sugar precursors (e.g., N-acetylmannosamine azide), which get metabolically integrated into sialylated N-glycans on RNAs.
Click chemistry enrichment: The azide tag enables a bioorthogonal reaction (e.g., DBCO-biotin), allowing selective capture of glycoRNAs using streptavidin or lectin beads.
2. RNA Isolation & Library Preparation
Enriched glycoRNAs are isolated via gentle extraction methods (TRIzol or column-based) to preserve RNA integrity and glycan linkages.
Library prep typically targets small non-coding RNAs (e.g., Y, snRNA, snoRNA), requiring modifications to standard adapters and reverse transcription protocols to accommodate glycan presence.
3. Sequencing Platforms & Readout
High-throughput platforms (Illumina SBS) are utilized for deep sequencing, optimized for low-abundance glycoRNAs.
Complementary technologies may include:
Northwestern blotting: Size-resolved validation of glycoRNAs.
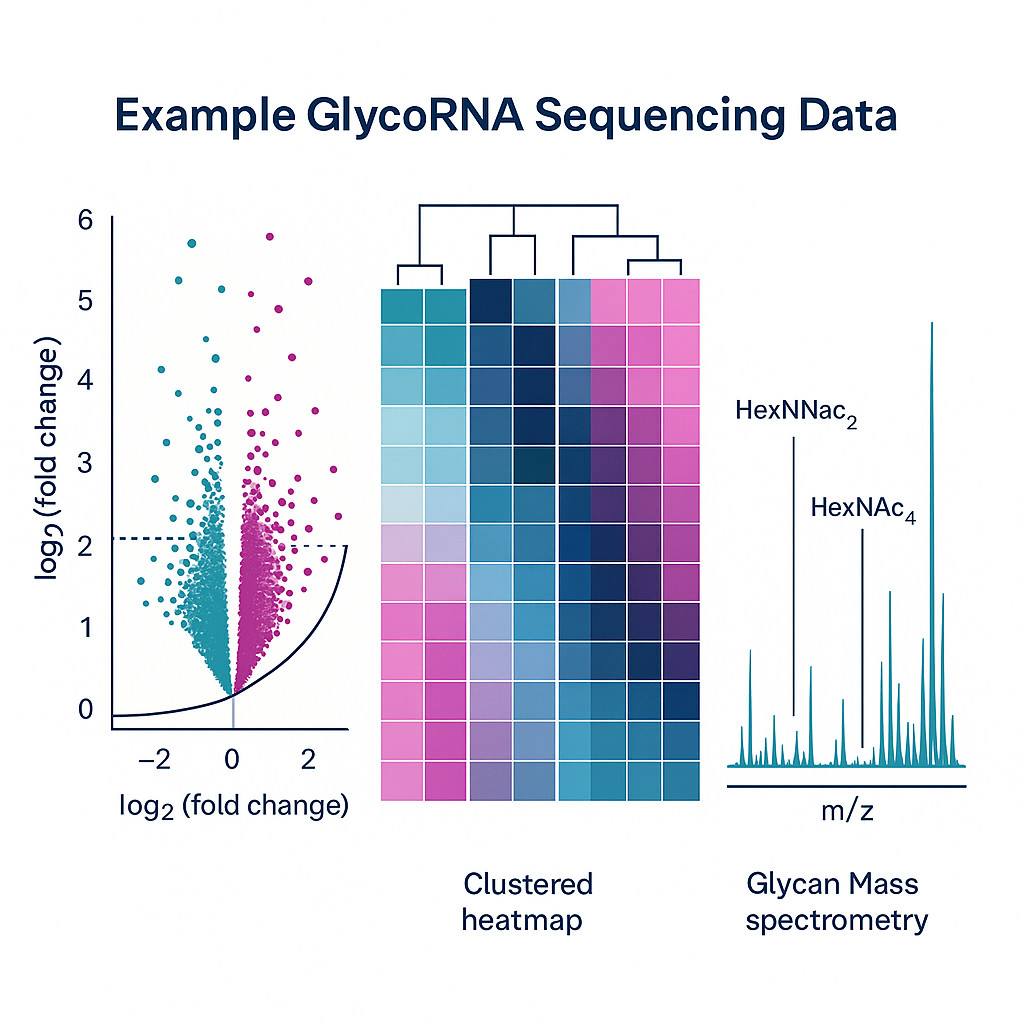
Mass spectrometry workflows like GlycanDIA: Characterize glycan structures—such as sialylated or fucosylated—and quantify glycoform diversity at depth.
Imaging (FRET, confocal): Spatially visualize glycoRNAs on cell membranes or extracellular vesicles using fluorescent tagging.
4. Data Analysis & Glyco-RNA Mapping
After sequencing, data pipelines analyze read abundance, sequence identity, and modification patterns.
Machine-learning tools (e.g., GlyinsRNA) predict glycosylation motifs with AUROC ≈ 0.79, helping identify likely glycosylation sites.
Glycan-RNA linkage is validated structurally with enzymatic assays (e.g., PNGase F sensitivity) and mass spectrometry.
Key Technical Challenges
Low glycoRNA abundance: GlycoRNAs are rare (~20 pmol/µg RNA), necessitating deep sequencing and sensitive enrichment.
Complex glycan structures: Structural heterogeneity demands robust MS methods like GlycanDIA .
Covalent linkage verification: Biochemical evidence—resistance to denaturation and sensitivity to PNGase F—confirms genuine glycan conjugation .
| Step |
Main Goal |
| Metabolic labeling + click |
Tag and isolate glycoRNAs |
| RNA extraction & library prep |
Preserve glycan-RNA integrity and generate cDNA |
| Sequencing + MS/imaging |
Identify glycoRNAs, define glycan profile, visualize |
| Bioinformatic analysis |
Map sequences, quantify, predict glycosylation motifs |
Applications of GlycoRNA Sequencing in Research and Pharma
1. Biomarker Discovery
- Cancer signatures: Studies have identified specific glycoRNAs that are enriched or depleted in breast cancer tissues versus healthy controls. These differential patterns suggest potential biomarkers for early detection and monitoring of disease progression.
- Extracellular vesicles: GlycoRNAs packaged into sEVs (exosomes) exhibit distinct glycosylation profiles in pathological states, offering minimally invasive biomarker sources.
2. Drug Target Validation & Mechanism Studies
- Regulatory glyco-miRNAs: Work in pancreatic cancer cells revealed that N-glycosylation of miR-21-5p affects tumor-related pathways like p53 signaling. Inhibiting glycosyltransferases (e.g., B4GALT1) altered tumor proliferation and apoptosis.
- Immune receptor interaction: GlycoRNAs bind to Siglec receptors (e.g., Siglec-11/14), implicating them in immune regulation and providing new avenues for immunomodulatory drug development.
3. Decoding Disease Mechanisms
- Neuroinflammation insights: Glycosylated Y-RNAs and snoRNAs in brain tissues may interact with RNA binding proteins and immune systems to influence neuroinflammatory responses.
- Cancer cell behavior: Imaging tools like ARPLA have mapped glycoRNAs within membrane lipid rafts, uncovering associations between glycoRNA abundance and tumor invasiveness—lower glycoRNA levels often correlate with metastasis.
4. Multi-omic Integration for Functional Context
Combining GlycoRNA sequencing with mass spectrometry, spatial imaging, and pathway analysis builds a holistic view:
- Elucidates glycan structures on specific RNA species.
- Assigns biological function through pathway enrichment (e.g., KEGG signaling in tumor growth) .
- Enables targeted validation using FRET, knockdown assays, and glycosyltransferase inhibition.
5. Pharmaceutical R&D & CRO Applications
By deploying our end-to-end GlycoRNA sequencing workflow, CROs and pharma teams can:
- Stratify patient subgroups based on glycoRNA signatures.
- Monitor drug response, especially in immuno-oncology, where glycan-RNA interactions modulate immune checkpoints.
- Identify off-target effects, leveraging glycoRNA changes as markers of cellular stress or toxicity.
Why Choose Our GlycoRNA Sequencing Service
✅ Best-in-Class Technical Platform
- Specialized enrichment methods – We use metabolic labeling (e.g., Ac4ManNAz) combined with click chemistry and lectin/streptavidin bead capture to selectively isolate glycoRNAs—a technique proven by leading providers in the field.
- Integrated, high-resolution readout – Sequencing is paired with mass spectrometry and glycoRNA imaging (e.g., FRET-based systems) for structural insights, mimicking the depth of established platforms .
✅ Accurate & Sensitive Profiling
- Low abundance detection – Our workflows are tuned to capture rare glycoRNA species via deep sequencing and sensitive MS, matching the categorization sensitivity of GlycoRNA-Seq services .
- Exact glycan resolution – Through tandem MS and glycan analysis (akin to GlycanDIA approaches), we map sialylation, fucosylation, and other glycoforms accurately.
✅ End-to-End, Customizable Workflow
- From sample to insight – Our platform supports cell lines, tissues, and biofluids (serum, sEVs) with minimal sample handling and high-quality preservation at every step including metabolic labeling, enrichment, sequencing, MS, imaging, and bioinformatics—mirroring commercial integrated solutions.
- Tailored protocols – Whether you're profiling extracellular vesicles, tissue biopsies, or subcellular fractions, protocols are optimized per project to ensure reliable results.
✅ Expert-Led Quality & Interpretation
- Seasoned scientific team – With experts in glycobiology and RNA biology guiding project design and data interpretation, our service parallels the expert-level guidance provided by top providers.
- Stringent QC at every stage – Quality control metrics such as enrichment specificity, glycoform detection rates, and sample integrity are monitored continuously to ensure publication-ready data.
✅ Fast Turnaround & Scalable Throughput
- Efficient pipelines – Using automated workflows and advanced platforms, we ensure rapid delivery of sequencing and MS results, similar to the high-throughput capacities noted in glycan sequencing providers.
- Flexible project sizing – From pilot studies to large-scale profiling, our services can be efficiently scaled to meet your research demands.
✅ Seamless Multi-Omic Integration
- Holistic insights – Combine glycoRNA sequencing with proteomics, metabolomics, and transcriptomics to develop a full molecular profile of your biological system.
- Spatial and structural context – Include optional, cutting-edge imaging (e.g., FRET-based glycoRNA visualization) and mass spectrometry analysis to map subcellular localization and glycoform diversity.
✅ Dedicated Customer Support
- Guided consultation – Our scientific liaisons help shape study design, choose the right methods, and outline data analysis frameworks.
- Expert report delivery – Receive interpretable, comprehensive reports covering glycoRNA identities, abundance, glycan structures, spatial distribution, and biological relevance.
Comparing Our Service vs. Traditional RNA-Seq
| Feature |
Our GlycoRNA Sequencing Service |
Traditional RNA-Seq |
| Target Molecule |
Glycosylated small ncRNAs |
Total RNA (mRNA, ncRNA) |
| Enrichment Strategy |
Metabolic labeling & click chemistry |
Poly(A) selection or ribodepletion |
| Glycan Structural Identification |
Yes (sialyl/fucosyl profiling via MS) |
No |
| Spatial & Interaction Insight |
Optional imaging (FRET, confocal) |
No |
| Ideal For |
Cell-surface glycoRNA studies, biomarker profiling |
Gene expression, transcriptome mapping |
✅ In Summary
Choosing our GlycoRNA sequencing service means gaining access to:
- A robust, targeted platform for detecting and profiling glycosylated RNAs.
- Sensitive and accurate measurement of rare RNA–glycan conjugates.
- Project customization and expert-level guidance at each stage.
- Fast and scalable throughput to meet diverse research needs.
- Integrated biological insights spanning sequencing, structure, and spatial mapping.
- Responsive support to help drive your project from conception to publication.
If you're aiming to explore the hidden landscape of RNA glycosylation with confidence, precision, and scientific rigor, our GlycoRNA sequencing service combines the latest technologies and expertise to deliver actionable, high-quality results.
How to Get Started: Sample Submission and Workflow
Step 1: Sample Submission
Accepted sample types:
- Cell lines (both treated and untreated),
- Tissue biopsies,
- Biofluids (serum, plasma),
- Small extracellular vesicles (sEVs).
- Minimum quantities required:
- ≥ 3 × 106 cultured cells (labeled),
- ≥ 25 μg total RNA (RIN ≥ 6),
- ≥ 200 μL serum/plasma,
- ≥ 1 × 1010 sEV particles.
Pre-labeling: For metabolic labeling, culture cells in the presence of azide-sugar precursors like Ac4ManNAz for ~48 hours before RNA extraction.
Sample handling
- Maintain RNase-free conditions (e.g., DEPC-treated consumables). For tissues and sEVs, snap freeze in liquid nitrogen and store at −80 °C. Avoid freeze–thaw cycles .
- Ship samples on dry ice with proper labeling, metadata, and chain-of-custody forms.
Step 2: Pre-Processing & Quality Control
Metabolic labeling / rPAL labeling
- Azide-sugar (Ac4ManNAz) incorporation: 48–72 hours in live cells.
- Alternatives: Periodate oxidation and aldehyde-biotin labeling (rPAL) for direct tagging of sialic-acid diols.
RNA extraction
- TRIzol or column-based protocols with proteinase K treatment to minimize glycoprotein contamination .
- QC metrics: RNA integrity (RIN ≥6), purity (260/280 ~2.0), and biotinylation efficiency via dot blot or fluorescence assay.
Step 3: Enrichment & Library Preparation
Selective glycoRNA enrichment:
- Click chemistry: React azide-modified glycans with DBCO-biotin; capture on streptavidin beads.
- rPAL method: Click biotin to periodate-oxidized glycoRNAs.
Library preparation:
- Custom small RNA library kits tailored for glycoRNA integrity, preserving glycan structures during reverse transcription and adapter ligation .
Step 4: Sequencing & Orthogonal Validation
- High-throughput sequencing (e.g., Illumina) performs deep coverage to detect low-abundance glycoRNAs.
- Mass spectrometry (LC-MS/MS or MALDI-TOF) resolves glycan structures—sialylation, fucosylation—and quantifies glycoform heterogeneity.
- Imaging options (optional): FRET-based spatial visualization of glycoRNAs in sEVs or live cells, ideal for localization and interaction studies.
Step 5: Data Analysis & Interpretation
Sequencing data processing:
- Adapter trimming, alignment (Bowtie/STAR), and quantification of glycoRNA species (snRNA, miRNA, Y-RNA, tRNA fragments).
Glycosylation mapping:
- Identify glyco-motifs, type of glycans, and site-specific modifications using motif search tools and MS data .
Integrated reporting:
- Differential abundance tables, glycoform profiles, and biological interpretation with GO/KEGG pathway insights aid hypothesis generation .
Interactive consultation:
- One-on-one session with our scientists to review results, address follow-up questions, and discuss validation or next steps.
Deliverables
- Raw sequencing reads (FASTQ), processed expression matrices, glycan annotation files, mass spectrometry spectra, imaging galleries (if requested), and an interpretive project report.
Related RNA Sequencing Services
To complement and extend your GlycoRNA research, we provide a range of advanced proteomics and glycomics services that integrate seamlessly with our GlycoRNA sequencing platform. These solutions enable a deeper understanding of glycosylation dynamics and protein–RNA interactions, helping you construct a complete molecular landscape.
By combining glycoRNA sequencing with these glycomics and multi-omic services, you gain a holistic view of cellular regulation—from molecular modifications to spatial organization and phenotypic consequences.
FAQs and Support
Can RNA actually be glycosylated?
Yes – the first definitive characterization of glycoRNAs occurred in 2021 (Flynn et al., Cell), where small non-coding RNAs (e.g., snRNA, snoRNA, tRNA, Y-RNA, miRNA) were found to carry covalent N-glycans, often with sialylated and fucosylated structures, on the cell surface.
What labeling strategies are used to enrich glycoRNAs?
We offer two validated enrichment approaches:
- Metabolic labeling + click chemistry – Using Ac4ManNAz, an azide-sugar precursor is metabolically incorporated into glycans. The azide group is then reacted with DBCO-biotin and captured on streptavidin beads.
- rPAL (periodate oxidation + aldehyde ligation) – Sialic acid moieties are selectively oxidized to aldehydes and conjugated to biotin without prior metabolic labeling.
These parallel strategies ensure high sensitivity and specificity for glycosylated RNAs.
What sequencing platforms and data output are provided?
- Next-generation sequencing (Illumina) for deep coverage of small RNAs.
- Standard bioinformatics including adapter trimming, alignment (Bowtie/STAR), and differential glycoRNA quantification.
- Mass spectrometry (LC-MS/MS or MALDI-TOF) to resolve glycan structures (e.g., sialylation, fucosylation).
- Optional imaging (e.g., drFRET or ARPLA) for spatial visualization in cells or extracellular vesicles.
- Deliverables include raw FASTQ files, processed glycoRNA tables, MS glycoform data, imaging galleries, and a comprehensive interpretive report.
How is specificity and accuracy ensured?
We apply rigorous controls and validation measures:
- Stringent bead-washing and negative labelling controls (no sugar/biotin).
- Enzymatic treatments (e.g., PNGase F or sialidase) to confirm glycan structures.
- Complementary biochemical tests like Clier-qPCR or Northern blotting for high-confidence identification.
What sample types and quantities are required?
Cells: ≥3 × 106 cultured cells with metabolic labeling.
Total RNA: ≥25 µg with RIN ≥ 6.
Biofluids: ≥200 µL serum/plasma or ≥1 × 1010 sEV particles.
Samples must be RNase-free, snap-frozen, and shipped on dry ice. Detailed instructions are provided on request.
Which support options are available?
- Consultation: Expert-led study design support.
- Data presentation: One-on-one report walkthrough and experimental follow-up suggestions.
- Technical support: Guidance for downstream validation (e.g., qPCR, imaging).
How do you control for contamination and false positives?
To ensure specificity, we implement multiple safeguards:
- Parallel negative controls (no metabolic label or no sugar precursor) processed alongside samples to detect background binding from oligo- or non-specific capture.
- Rigorous RNase-negative clean lab procedures, and inclusion of blank libraries, to prevent environmental nucleic acid contamination—a critical consideration in low-input sequencing .
- Enzymatic validation: PNGase F and sialidase treatments are used to confirm removal of glycan moieties and loss of signal only occurs when glycosylated structures are present.
Are glycoRNA levels comparable across different cell types?
Not always—metabolic labeling efficiency can vary:
- Some cell types incorporate azide-sugar precursors (like Ac4ManNAz) much less efficiently, leading to strong differences in labeling intensity between samples.
- We recommend pilot studies and normalization strategies—such as ratio-based quantification across matched controls—to account for variable labeling efficiencies and ensure meaningful cross-sample comparison.