In the intricate theater of cellular biology, protein phosphorylation and dephosphorylation play pivotal, yet often overlooked roles. These seemingly simple chemical reactions form the cornerstone of complex regulatory networks within cells. Acting as molecular switches, they can instantaneously alter protein function, activity, and interactions, thereby influencing a myriad of biological processes ranging from cell growth to gene expression.
What Are Phosphorylation and Dephosphorylation:Molecular On/Off Switches
Phosphorylation involves the addition of a phosphate group (PO₄³⁻) to specific amino acid residues on proteins, typically serine, threonine, or tyrosine . This process is catalyzed by kinases and generally requires ATP as a phosphate donor. Conversely, dephosphorylation, catalyzed by phosphatases, removes phosphate groups from proteins .The delicate balance between these processes regulates protein "on" and "off" states:
| Process | Catalyst | Action | Biological Effect |
|---|---|---|---|
| Phosphorylation | Kinases | Add phosphate group | Activate or inhibit proteins |
| Dephosphorylation | Phosphatases | Remove phosphate group | Reverse phosphorylation effects |
Key Enzymes: Kinases and Phosphatases
Protein kinases and phosphatases are pivotal enzymatic regulators of protein phosphorylation states. The human genome encodes over 500 kinases and approximately 200 phosphatases, reflecting an asymmetric balance that is conserved across diverse species from yeast to humans. This numerical disparity underscores the intricate nature of phosphorylation-based signaling networks.
Kinases exhibit heightened sensitivity to cellular perturbations and are more prone to phosphorylation themselves. A comprehensive analysis of over 1,400 perturbation experiments in yeast revealed differential expression in about 34% of kinases, a significantly higher proportion compared to phosphatases . Moreover, kinases demonstrate a broader spectrum of phenotypic impacts when manipulated, highlighting their versatility in cellular responses.
In contrast, phosphatases appear to be more stable entities. They are less susceptible to phosphorylation, with studies indicating that while over 40% of kinases possess phosphorylation capacity, this figure drops to at most 12% for phosphatases . This stark difference in responsiveness and regulatory mechanisms between kinases and phosphatases contributes to the fine-tuning of cellular signaling pathways.
The spatial distribution of kinases and phosphatases within cells plays a crucial role in regulating protein phosphorylation. This compartmentalization can generate gradients of phosphorylated substrates across various subcellular domains, influencing a myriad of cellular processes. Key signaling cascades such as MAPK, PI3K/Akt/mTOR, PKA, and PKC interact at multiple intracellular loci, further emphasizing the significance of the spatial organization of these enzymatic regulators.
Dynamic Balance Between Phosphorylation and Dephosphorylation
The interplay between phosphorylation and dephosphorylation indeed serves as a crucial molecular switch in cellular regulation, playing a vital role in maintaining proper cellular function and responding to various stimuli. This dynamic balance is essential for numerous physiological processes and can have significant consequences when disrupted.
Phosphorylation and dephosphorylation events are mediated by kinases and phosphatases, respectively, creating a constant equilibrium that allows for rapid and reversible changes in protein function, activity, and interactions. This molecular switch mechanism enables cells to respond quickly to external stimuli and maintain homeostasis.
The importance of this balance is evident in several key cellular processes:
Cell signaling: Phosphorylation/dephosphorylation cycles regulate signaling pathways, controlling cell survival, growth, and differentiation.
Protein activation/deactivation: For example, protein kinase B is activated by phosphorylation of its Ser and Thr residues, regulating cell survival.
Subcellular localization: Phosphorylation states can influence protein localization within the cell, affecting their function.
Enzyme regulation: The cyclic interconversion between phosphorylated and unphosphorylated forms of enzymes is a major mechanism of cellular regulation.
When the balance between phosphorylation and dephosphorylation is disrupted, it can lead to severe consequences:
Cancer: Abnormal phosphorylation patterns are often associated with oncogenic signaling and tumor progression.
Diabetes: Dysregulation of phosphorylation-dependent pathways can contribute to insulin resistance and metabolic disorders.
Neurodegenerative disorders: Imbalances in protein phosphorylation, such as hyperphosphorylation of tau in Alzheimer's disease, are linked to neuronal dysfunction.
The spatial and temporal control of phosphorylation/dephosphorylation cycles is crucial for maintaining cellular homeostasis. This control is influenced by factors such as enzyme localization, formation of multiprotein complexes, and the local lipid environment of membrane proteins.
Recent research has highlighted the complexity of these molecular switches. For instance, studies have shown that phosphorylation can regulate protein function through various mechanisms, including conformational changes and creation of binding sites for other proteins. Additionally, some kinases, such as Akt/PKB, are regulated by a disorder-to-order transition of their activation loop upon phosphorylation.
Understanding the intricacies of this molecular switch mechanism is vital for developing targeted therapies and interventions for various diseases associated with disrupted phosphorylation balance. As our knowledge of these processes deepens, it opens up new avenues for therapeutic approaches in treating complex disorders like cancer, diabetes, and neurodegenerative diseases.
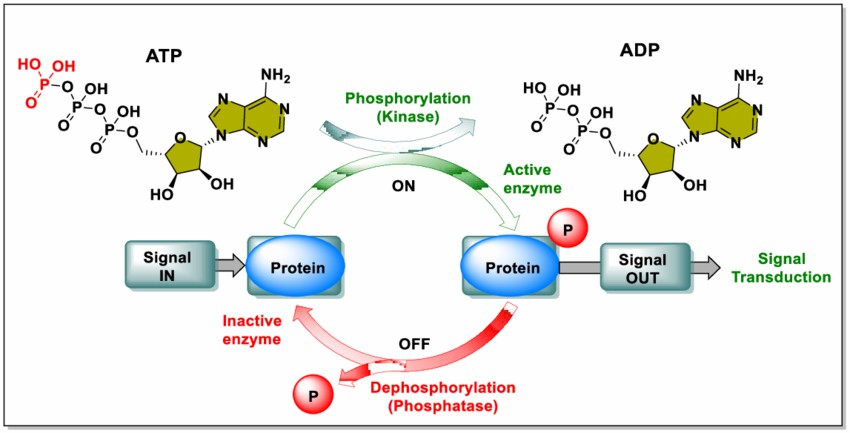 Fig 1: Schematic diagram of phosphorylation and dephosphorylation. (Vidya Jyothi Alli et al,. 2023)
Fig 1: Schematic diagram of phosphorylation and dephosphorylation. (Vidya Jyothi Alli et al,. 2023)
Comprehensive Comparison of Phosphorylation and Dephosphorylation
| Aspect | Phosphorylation | Dephosphorylation | Examples |
|---|---|---|---|
| Definition | Addition of a phosphate group (PO₄³⁻) to a protein or molecule. | Removal of a phosphate group (PO₄³⁻) from a protein or molecule. | Phosphorylation of CREB (cAMP response element-binding protein) in neurons enhances its ability to activate transcription. |
| Enzyme Involved | Kinases | Phosphatases | Protein kinase A (PKA) phosphorylates numerous substrates, while protein phosphatase 1 (PP1) dephosphorylates many of the same targets. |
| Energy Requirement | Requires ATP as a phosphate donor. | Does not require ATP; uses water in a hydrolysis reaction. | The phosphorylation of glucose by hexokinase in glycolysis requires ATP. |
| Common Target Residues | Serine, threonine, tyrosine in eukaryotes. | Same residues as phosphorylation. | Insulin receptor substrate-1 (IRS-1) is phosphorylated on multiple serine and threonine residues. |
| Biological Function | Activates or deactivates proteins, regulates signaling pathways. | Reverses phosphorylation effects, restoring protein to its original state. | Phosphorylation of glycogen synthase kinase-3 (GSK-3) by Akt inhibits its activity, promoting glycogen synthesis. |
| Cellular Role | Facilitates protein-protein interactions, alters activity or localization. | Terminates or modulates signaling events, maintains homeostasis. | Phosphorylation of SNAP25 reduces its binding affinity for syntaxin-1A, altering its cellular location. |
| Reversibility | Reversible process as it pairs with dephosphorylation. | Reversible process as it pairs with phosphorylation. | The reversible phosphorylation of Na⁺/K⁺-ATPase regulates its activity in ion transport across cell membranes. |
| Examples in Pathways | MAPK/ERK pathway, PI3K/Akt signaling. | Inactivation of kinases or termination of cellular signals. | ERK1/2 phosphorylation in the MAPK pathway promotes cell proliferation, while its dephosphorylation by DUSP6 attenuates signaling. |
| Detection Methods | Mass spectrometry, Western blotting, phospho-specific antibodies. | Indirect methods by detecting absence of phosphate groups. | Phosphoproteomics using mass spectrometry can identify thousands of phosphorylation sites in a single experiment. |
| Impact on Diseases | Abnormal phosphorylation is linked to cancer, diabetes, and Alzheimer's. | Impaired dephosphorylation is associated with hyperphosphorylated tau in Alzheimer's. | Hyperphosphorylation of tau protein is a hallmark of Alzheimer's disease, resulting from an imbalance between kinase and phosphatase activities. |
Phosphorylation and Dephosphorylation in Disease
Phosphorylation and dephosphorylation are fundamental processes that regulate cellular functions, but when these processes are disrupted, they can contribute to a range of diseases. Understanding how dysregulated phosphorylation or dephosphorylation affects cellular processes is crucial for developing targeted therapies. Below, we explore examples of diseases linked to phosphorylation imbalances, the role of overactive kinases or underactive phosphatases, and potential therapeutic targets for disease treatment.
Examples of Diseases Linked to Dysregulated Phosphorylation or Dephosphorylation
Cancer:
Dysregulation of phosphorylation plays a central role in many cancers. For instance, the overexpression and constitutive activation of EGFR in cancer cells lead to abnormal phosphorylation patterns, promoting tumor growth and progression. In glioblastoma, EGFR gene amplification and increased EGFR expression are detected in 40-50% of cases.
Neurodegenerative Diseases:
In Alzheimer's disease, tau protein becomes abnormally hyperphosphorylated, leading to the formation of neurofibrillary tangles. The hyperphosphorylation of tau occurs to a stoichiometry at least three-fold greater than normal tau, contributing to the aggregation of paired helical filaments.
The Role of Overactive Kinases or Underactive Phosphatases in Diseases
Overactive Kinases:
In cancer, overactive kinases can lead to excessive cell proliferation and survival. For example, mutations in the BRAF kinase are found in approximately 50% of melanomas, leading to constitutive activation of the MAPK pathway.
Underactive Phosphatases:
The underactivity of phosphatases can contribute to disease progression. For instance, loss of function of the tumor suppressor PTEN, a lipid phosphatase, is frequently observed in various cancers and leads to hyperactivation of the PI3K/AKT pathway.
How Phosphorylation/Dephosphorylation Imbalances Contribute to Disease Progression
Cancer Progression:
Imbalanced phosphorylation can promote cancer development by altering key pathways. In a study of human cancers, an atlas of the human kinome revealed how tumors systematically hijack particular subnetworks through aberrant phosphorylation.
Neurodegeneration:
In Alzheimer's disease, the abnormal hyperphosphorylation of tau leads to the formation of neurofibrillary tangles, a histopathological hallmark of the disease. This process is crucial to the progression of neurofibrillary degeneration.
Potential Therapeutic Targets Related to Modulating Kinase or Phosphatase Activity
Kinase Inhibitors:
Targeting overactive kinases with specific inhibitors is a promising approach in cancer therapy. For example, EGFR inhibitors have been developed to target the dysregulated EGFR signaling in various cancers.
Phosphatase Modulators:
Modulating phosphatase activity can restore normal signaling. Research has shown that pathological tau from Alzheimer's brain can induce site-specific hyperphosphorylation of tau, suggesting that targeting tau phosphorylation could be a potential therapeutic approach.
Explore how phosphorylation analysis supports drug discovery in this article: What is Protein Phosphorylation?.
Techniques for Studying Protein Phosphorylation and Dephosphorylation
Researchers use several techniques to investigate these modifications, including:
Western Blotting: Detects phosphorylated proteins using phospho-specific antibodies. For example, a study by Ardito et al. demonstrated the use of western blotting to detect phosphorylation of ERK1/2 in response to EGF stimulation in cancer cells.
Mass Spectrometry: A cornerstone of phosphoproteomics, enabling large-scale identification and quantification of phosphorylation sites. Creixell et al. used mass spectrometry-based phosphoproteomics to create an atlas of the human kinome, revealing how tumors systematically hijack particular subnetworks through aberrant phosphorylation.
Electrophoretic Mobility Shift Assays: Identify phosphorylation-induced changes in protein mobility. This technique was used by Gong et al. to study the hyperphosphorylation of tau protein in Alzheimer's disease, showing that phosphorylation can alter the electrophoretic mobility of proteins.
For an in-depth summary, visit our resource: Protein Phosphorylation Detection Methods.
signaling and glucose homeostasis.
Phosphoproteomics: Large-Scale Analysis of Protein Phosphorylation
Mass spectrometry-based phosphoproteomics is transforming how scientists study protein phosphorylation. Key steps include:
- Protein Extraction: Harvesting proteins from cells or tissues.
- Enzymatic Digestion: Breaking down proteins into peptides.
- Phosphopeptide Enrichment: Isolating phosphorylated peptides.
- Mass Spectrometric Analysis: High-throughput identification of phosphorylation sites.
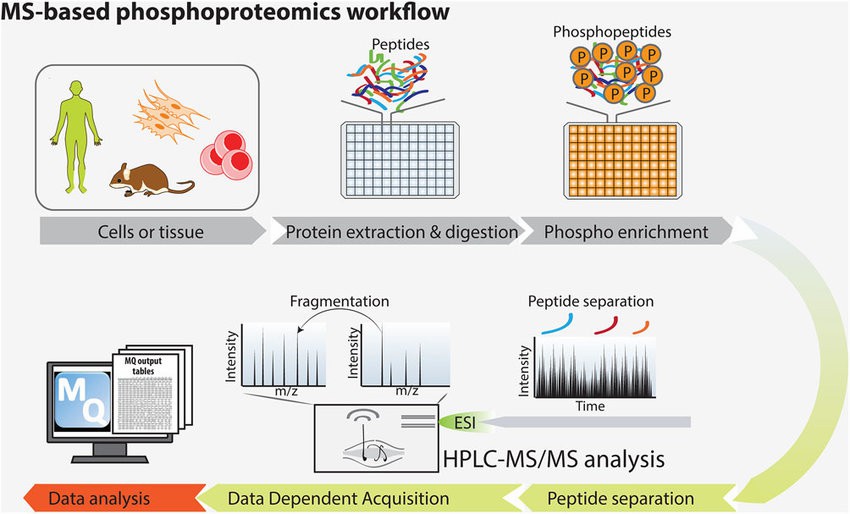 Figure 2: The basic principle of the phosphoproteomics approach. (Francesca Sacco et al,. 2015)
Figure 2: The basic principle of the phosphoproteomics approach. (Francesca Sacco et al,. 2015)
Select Service
Learn more about advancements in this field through our dedicated resource: Advancements in Protein Phosphorylation Research.
Challenges in Phosphorylation Research
Phosphorylation research indeed faces significant challenges due to the low abundance of phosphorylated proteins, weak ionization of phosphopeptides during mass spectrometry, and the transient nature of phosphorylation events. These challenges have been well-documented in scientific literature, and researchers have developed various strategies to address them.
Low Abundance of Phosphorylated Proteins
The low abundance of phosphorylated proteins poses a major challenge in phosphorylation research. This issue is particularly problematic when studying weakly phosphorylated or low-abundance proteins. To overcome this challenge, researchers have developed several strategies:
Immunoprecipitation: Using antibodies to concentrate and load more protein in sample lanes for Western blot analysis.
Enrichment techniques: Implementing advanced fractionation and labeling techniques to promote more efficient mass spectrometry (MS) analysis of phosphoproteins.
Knock-in models: Developing knock-in mouse models with tagged proteins of interest to facilitate immuno-affinity purification and subsequent MS analysis.
Weak Ionization of Phosphopeptides
Phosphopeptides often exhibit weak ionization during mass spectrometry, which can hinder their detection and quantification. Researchers have addressed this issue through various approaches:
Collision-induced dissociation (CID): This technique has proven effective for phosphorylation site identification and assignment, although it can lead to the loss of phosphate groups.
Optimized MS acquisition strategies: Implementing appropriate MS acquisition methods is critical for identifying and quantifying protein phosphorylation, often more so than sample-specific enrichment or labeling approaches.
Phosphatase treatment: Comparing phosphatase-treated samples with mock-treated samples to determine fractional phosphorylation occupancy.
Transient Nature of Phosphorylation Events
The temporal component of phosphorylation events presents another significant challenge in phosphorylation research. Strategies to address this issue include:
Kinase and phosphatase inhibitors: Adding these inhibitors during sample preparation to minimize extraneous phosphorylation events and reduce sample-to-sample variability.
Targeted hypothesis-driven investigations: Focusing on specific, known phosphorylation sites of interest rather than conducting broad discovery-based analyses.
Quantitative MS analysis: Using ion intensity ratios of phosphorylated peptides versus corresponding unphosphorylated peptides to determine phosphorylation levels.
These challenges and strategies highlight the complexity of phosphorylation research and the ongoing efforts to improve detection and quantification methods. As technologies continue to advance, researchers are developing more sensitive and accurate techniques to study protein phosphorylation and its biological significance.
Our advanced PTMs Proteomics Services overcome these hurdles, delivering reliable and accurate results for your research needs. Visit: Post-Translational Modifications Service.
Emerging Trends in Phosphorylation Research
Cutting-edge advancements are paving the way for deeper insights into phosphorylation:
- Single-Cell Phosphoproteomics: Enables study of phosphorylation at the cellular level.
- Integration with Other -Omics Data: Links phosphorylation with genomics, transcriptomics, and metabolomics.
- Non-Canonical Phosphorylation Sites: Uncovers new mechanisms and functions.
- AI-Driven Analysis: Facilitates prediction of phosphorylation sites and patterns.
Transform Your Research with Creative Proteomics
Phosphorylation and dephosphorylation are at the heart of cellular regulation, impacting everything from signaling to disease progression. Understanding these modifications is critical for advancing research and developing new therapies.
With our cutting-edge services and expert team, Creative Proteomics is here to support your research endeavors. Contact us today to unlock the full potential of phosphorylation analysis in your projects. Together, let's drive innovation in molecular biology and drug discovery.
Our commitment to quality and precision makes us a trusted partner for academic and industrial researchers alike.
At Creative Proteomics, we provide tailored solutions for your phosphorylation research, including:
- Comprehensive phosphoproteomics services.
- Expertise in phosphosite mapping.
- Advanced quantitative analyses of phosphorylated proteins.
People Also Ask
1. What is the difference between phosphorylation and dephosphorylation?
Phosphorylation involves the addition of a phosphate group to a protein, typically catalyzed by kinases. Dephosphorylation is the removal of a phosphate group, catalyzed by phosphatases. These processes are reversible and play crucial roles in regulating protein function.
2. How common is protein phosphorylation?
Protein phosphorylation is extremely common, with approximately 13,000 human proteins having sites that can be phosphorylated.
3. What techniques are used to study protein phosphorylation?
Common techniques include Western blotting, mass spectrometry, phospho-specific antibodies, and phosphoproteomics. Each method offers unique advantages for detecting and quantifying phosphorylation events
4. How does phosphorylation affect protein function?
Phosphorylation can activate or deactivate proteins, alter their binding affinities, change their subcellular localization, or modify their interactions with other molecules. These changes can significantly impact cellular signaling and regulation.
References
- Lonze, B. E., & Ginty, D. D. (2002). Function and regulation of CREB family transcription factors in the nervous system. Neuron, 35(4), 605-623. https://doi.org/10.1016/S0896-6273(02)00828-0
- Cohen, P. T. (2002). Protein phosphatase 1 – targeted in many directions. Journal of Cell Science, 115(2), 241-256. https://doi.org/10.1242/jcs.115.2.241
- Wilson, J. E. (2003). Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. Journal of Experimental Biology, 206(12), 2049-2057. https://doi.org/10.1242/jeb.00241
- Gual, P., Le Marchand-Brustel, Y., & Tanti, J. F. (2005). Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie, 87(1), 99-109. https://doi.org/10.1016/j.biochi.2004.10.019
- Cross, D. A., Alessi, D. R., Cohen, P., Andjelkovich, M., & Hemmings, B. A. (1995). Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature, 378(6559), 785-789. https://doi.org/10.1038/378785a0
- Rickman, C., & Duncan, R. R. (2010). Munc18/Syntaxin interaction kinetics control secretory vesicle dynamics. Journal of Biological Chemistry, 285(6), 3965-3972. https://doi.org/10.1074/jbc.M109.040402
- Poulsen, H., Morth, P., Egebjerg, J., & Nissen, P. (2010). Phosphorylation of the Na+,K+-ATPase and the H+,K+-ATPase. FEBS Letters, 584(12), 2589-2595. https://doi.org/10.1016/j.febslet.2010.04.035
- Caunt, C. J., & Keyse, S. M. (2013). Dual-specificity MAP kinase phosphatases (MKPs): shaping the outcome of MAP kinase signalling. FEBS Journal, 280(2), 489-504. https://doi.org/10.1111/j.1742-4658.2012.08716.x
- Humphrey, S. J., James, D. E., & Mann, M. (2015). Protein phosphorylation: a major switch mechanism for metabolic regulation. Trends in Endocrinology & Metabolism, 26(12), 676-687. https://doi.org/10.1016/j.tem.2015.09.013
- Iqbal, K., Liu, F., Gong, C. X., & Grundke-Iqbal, I. (2010). Tau in Alzheimer disease and related tauopathies. Current Alzheimer Research, 7(8), 656-664. https://doi.org/10.2174/156720510793611592
- Ardito, F., Giuliani, M., Perrone, D., Troiano, G., & Lo Muzio, L. (2017). The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. International Journal of Molecular Medicine, 40(2), 271-280. https://doi.org/10.3892/ijmm.2017.3036
- Bhaskaran, S., Hsu, T. C., Engel, M., Hanna-Rose, W., & Jankowsky, E. (2017). An Asymmetrically Balanced Organization of Kinases versus Phosphatases across Eukaryotes Determines Their Distinct Impacts. PLOS Computational Biology, 13(1), e1005221. https://doi.org/10.1371/journal.pcbi.1005221
- Bononi, A., Agnoletto, C., De Marchi, E., Marchi, S., Patergnani, S., Bonora, M., Giorgi, C., Missiroli, S., Poletti, F., Rimessi, A., & Pinton, P. (2011). Protein Kinases and Phosphatases in the Control of Cell Fate. Enzyme Research, 2011, 329098. https://doi.org/10.4061/2011/329098
- Guo, G., Gong, K., Wohlfeld, B., Hatanpaa, K. J., Zhao, D., & Habib, A. A. (2015). Ligand-Independent EGFR Signaling. Cancer Research, 75(17), 3436-3441. https://doi.org/10.1158/0008-5472.CAN-15-0989
- Gong, C. X., Liu, F., Grundke-Iqbal, I., & Iqbal, K. (2005). Post-translational modifications of tau protein in Alzheimer's disease. Journal of Neural Transmission, 112(6), 813-838. https://doi.org/10.1007/s00702-004-0221-0
- Creixell, P., Schoof, E. M., Simpson, C. D., Longden, J., Miller, C. J., Lou, H. J., ... & Linding, R. (2015). Kinome-wide decoding of network-attacking mutations rewiring cancer signaling. Cell, 163(1), 202-217. https://doi.org/10.1016/j.cell.2015.08.056
- Brand, T. M., Iida, M., & Wheeler, D. L. (2011). Molecular mechanisms of resistance to the EGFR monoclonal antibody cetuximab. Cancer Biology & Therapy, 11(9), 777-792. https://doi.org/10.4161/cbt.11.9.15050
- Nita-Lazar, A., Saito-Benz, H., & White, F. M. (2008). Quantitative phosphoproteomics by mass spectrometry: Past, present, and future. Proteomics, 8(21), 4433-4443. https://doi.org/10.1002/pmic.200800231
- Bio-Techne. (2024). Western Blot for Phosphorylated Proteins - Tips & Troubleshooting. https://www.bio-techne.com/resources/blogs/tips-to-optimize-your-western-blot-for-phosphorylated-protein-detection
- Rigbolt, K. T., & Blagoev, B. (2012). Quantitative phosphoproteomics to characterize signaling networks. Seminars in Cell & Developmental Biology, 23(8), 863-871. https://doi.org/10.1016/j.semcdb.2012.05.006
- Ito, T., Hoshino, T., Hishinuma, S., Nakata, M., Naruse, H., Yoshida, M., Sakamoto, A., Tanaka, K., Masukawa, D., Goto, K., & Inoue, R. (2017). Strategy for Identification of Phosphorylation Levels of Low Abundance Proteins in Vivo. Journal of Proteome Research, 16(12), 4453-4465. https://doi.org/10.1021/acs.jproteome.7b00561
Our products and services are for research use only.

